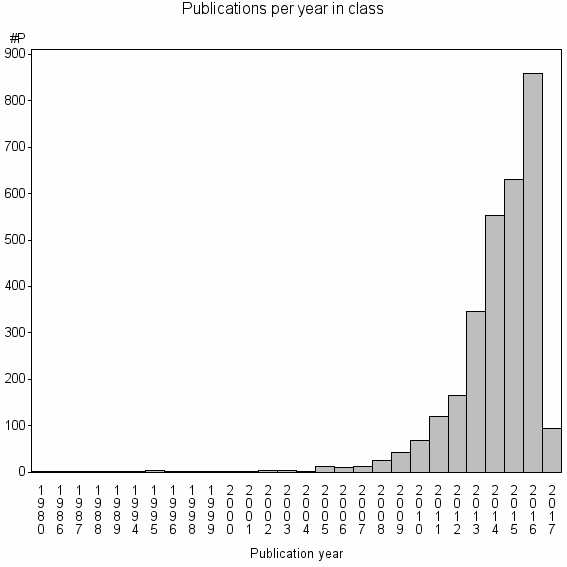
Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
| 915 | 2962 | 51.0 | 93% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 7 | 4 | INFECTIOUS DISEASES//MICROBIOLOGY//VIROLOGY | 1353914 |
| 33 | 3 | JOURNAL OF BACTERIOLOGY//MICROBIOLOGY//MOLECULAR MICROBIOLOGY | 107033 |
| 1115 | 2 | GENOME EDITING//CRISPR CAS9//CRISPR | 9706 |
| 915 | 1 | GENOME EDITING//CRISPR//CRISPR CAS9 | 2962 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | GENOME EDITING | authKW | 1842553 | 9% | 65% | 275 |
| 2 | CRISPR | authKW | 1703633 | 9% | 61% | 270 |
| 3 | CRISPR CAS9 | authKW | 1696592 | 9% | 65% | 255 |
| 4 | CAS9 | authKW | 1307140 | 5% | 82% | 154 |
| 5 | TALEN | authKW | 1073031 | 5% | 69% | 151 |
| 6 | ZINC FINGER NUCLEASE | authKW | 716124 | 4% | 59% | 117 |
| 7 | CRISPR CAS | authKW | 700126 | 3% | 77% | 88 |
| 8 | GENE EDITING | authKW | 533022 | 3% | 60% | 86 |
| 9 | GENOME ENGINEERING | authKW | 487036 | 3% | 49% | 96 |
| 10 | TRANSCRIPTION ACTIVATOR LIKE EFFECTOR NUCLEASES | authKW | 350871 | 1% | 85% | 40 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Biotechnology & Applied Microbiology | 11117 | 27% | 0% | 788 |
| 2 | Biochemistry & Molecular Biology | 7594 | 43% | 0% | 1277 |
| 3 | Genetics & Heredity | 6413 | 20% | 0% | 599 |
| 4 | Cell Biology | 3866 | 20% | 0% | 588 |
| 5 | Cell & Tissue Engineering | 3010 | 3% | 0% | 90 |
| 6 | Biochemical Research Methods | 2773 | 11% | 0% | 325 |
| 7 | Developmental Biology | 825 | 4% | 0% | 110 |
| 8 | Microbiology | 761 | 8% | 0% | 239 |
| 9 | Medicine, Research & Experimental | 691 | 7% | 0% | 202 |
| 10 | Plant Sciences | 384 | 7% | 0% | 195 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | GENOME ENGN | 313497 | 3% | 37% | 82 |
| 2 | INNOVAT GENOM INITIAT | 164899 | 1% | 67% | 24 |
| 3 | REGULAT INFECT BIOL | 118831 | 0% | 82% | 14 |
| 4 | CREAT INITIAT GENOME ENGN | 116124 | 0% | 87% | 13 |
| 5 | MOL PATHOL UNIT | 71982 | 2% | 13% | 54 |
| 6 | RNA SYST BIOL | 59949 | 1% | 36% | 16 |
| 7 | COMPUTAT INTEGRAT BIOL | 58730 | 2% | 10% | 55 |
| 8 | OURCE MUTANT MICE | 49460 | 0% | 40% | 12 |
| 9 | COMBINED PROGRAM MICROBIOL IMMUNOL | 48370 | 0% | 36% | 13 |
| 10 | ARCHAEA | 46828 | 1% | 22% | 21 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | NATURE BIOTECHNOLOGY | 25071 | 3% | 3% | 80 |
| 2 | NATURE METHODS | 17680 | 2% | 3% | 54 |
| 3 | RNA BIOLOGY | 13702 | 1% | 4% | 35 |
| 4 | JOURNAL OF GENETICS AND GENOMICS | 10852 | 1% | 4% | 25 |
| 5 | MOLECULAR THERAPY-NUCLEIC ACIDS | 9831 | 1% | 6% | 17 |
| 6 | NUCLEIC ACIDS RESEARCH | 7590 | 6% | 0% | 173 |
| 7 | SCIENTIFIC REPORTS | 7095 | 6% | 0% | 175 |
| 8 | ACS SYNTHETIC BIOLOGY | 6644 | 1% | 3% | 19 |
| 9 | G3-GENES GENOMES GENETICS | 5940 | 1% | 2% | 28 |
| 10 | MOLECULAR THERAPY | 5814 | 2% | 1% | 45 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | GENOME EDITING | 1842553 | 9% | 65% | 275 | Search GENOME+EDITING | Search GENOME+EDITING |
| 2 | CRISPR | 1703633 | 9% | 61% | 270 | Search CRISPR | Search CRISPR |
| 3 | CRISPR CAS9 | 1696592 | 9% | 65% | 255 | Search CRISPR+CAS9 | Search CRISPR+CAS9 |
| 4 | CAS9 | 1307140 | 5% | 82% | 154 | Search CAS9 | Search CAS9 |
| 5 | TALEN | 1073031 | 5% | 69% | 151 | Search TALEN | Search TALEN |
| 6 | ZINC FINGER NUCLEASE | 716124 | 4% | 59% | 117 | Search ZINC+FINGER+NUCLEASE | Search ZINC+FINGER+NUCLEASE |
| 7 | CRISPR CAS | 700126 | 3% | 77% | 88 | Search CRISPR+CAS | Search CRISPR+CAS |
| 8 | GENE EDITING | 533022 | 3% | 60% | 86 | Search GENE+EDITING | Search GENE+EDITING |
| 9 | GENOME ENGINEERING | 487036 | 3% | 49% | 96 | Search GENOME+ENGINEERING | Search GENOME+ENGINEERING |
| 10 | TRANSCRIPTION ACTIVATOR LIKE EFFECTOR NUCLEASES | 350871 | 1% | 85% | 40 | Search TRANSCRIPTION+ACTIVATOR+LIKE+EFFECTOR+NUCLEASES | Search TRANSCRIPTION+ACTIVATOR+LIKE+EFFECTOR+NUCLEASES |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | WRIGHT, AV , NUNEZ, JK , DOUDNA, JA , (2016) BIOLOGY AND APPLICATIONS OF CRISPR SYSTEMS: HARNESSING NATURE'S TOOLBOX FOR GENOME ENGINEERING.CELL. VOL. 164. ISSUE 1-2. P. 29 -44 | 137 | 93% | 74 |
| 2 | DOUDNA, JA , CHARPENTIER, E , (2014) THE NEW FRONTIER OF GENOME ENGINEERING WITH CRISPR-CAS9.SCIENCE. VOL. 346. ISSUE 6213. P. 1077 -+ | 119 | 82% | 552 |
| 3 | MAEDER, ML , GERSBACH, CA , (2016) GENOME-EDITING TECHNOLOGIES FOR GENE AND CELL THERAPY.MOLECULAR THERAPY. VOL. 24. ISSUE 3. P. 430 -446 | 185 | 65% | 35 |
| 4 | KIM, H , KIM, JS , (2014) A GUIDE TO GENOME ENGINEERING WITH PROGRAMMABLE NUCLEASES.NATURE REVIEWS GENETICS. VOL. 15. ISSUE 5. P. 321 -334 | 150 | 88% | 246 |
| 5 | CARROLL, D , (2014) GENOME ENGINEERING WITH TARGETABLE NUCLEASES.ANNUAL REVIEW OF BIOCHEMISTRY, VOL 83. VOL. 83. ISSUE . P. 409 -439 | 200 | 78% | 121 |
| 6 | HSU, PD , LANDER, ES , ZHANG, F , (2014) DEVELOPMENT AND APPLICATIONS OF CRISPR-CAS9 FOR GENOME ENGINEERING.CELL. VOL. 157. ISSUE 6. P. 1262 -1278 | 78 | 86% | 758 |
| 7 | MOHANRAJU, P , MAKAROVA, KS , ZETSCHE, B , ZHANG, F , KOONIN, EV , VAN DER OOST, J , (2016) DIVERSE EVOLUTIONARY ROOTS AND MECHANISTIC VARIATIONS OF THE CRISPR-CAS SYSTEMS.SCIENCE. VOL. 353. ISSUE 6299. P. - | 153 | 96% | 8 |
| 8 | GAJ, T , GERSBACH, CA , BARBAS, CF , (2013) ZFN, TALEN, AND CRISPR/CAS-BASED METHODS FOR GENOME ENGINEERING.TRENDS IN BIOTECHNOLOGY. VOL. 31. ISSUE 7. P. 397-405 | 89 | 77% | 805 |
| 9 | SANDER, JD , JOUNG, JK , (2014) CRISPR-CAS SYSTEMS FOR EDITING, REGULATING AND TARGETING GENOMES.NATURE BIOTECHNOLOGY. VOL. 32. ISSUE 4. P. 347 -355 | 86 | 95% | 337 |
| 10 | MAKAROVA, KS , WOLF, YI , ALKHNBASHI, OS , COSTA, F , SHAH, SA , SAUNDERS, SJ , BARRANGOU, R , BROUNS, SJJ , CHARPENTIER, E , HAFT, DH , ET AL (2015) AN UPDATED EVOLUTIONARY CLASSIFICATION OF CRISPR-CAS SYSTEMS.NATURE REVIEWS MICROBIOLOGY. VOL. 13. ISSUE 11. P. 722 -736 | 103 | 91% | 112 |
Classes with closest relation at Level 1 |