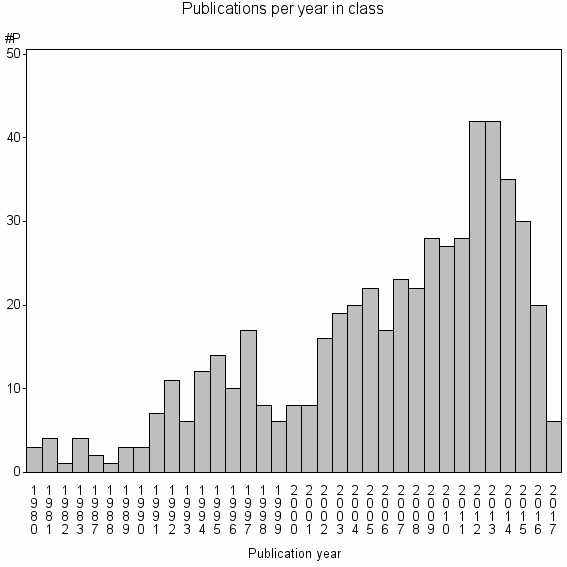
Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
| 19037 | 525 | 43.5 | 81% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 5 | 4 | CHEMISTRY, ORGANIC//CHEMISTRY, INORGANIC & NUCLEAR//CHEMISTRY, MULTIDISCIPLINARY | 1745167 |
| 189 | 3 | JOURNAL OF MAGNETIC RESONANCE//MAGNETIC RESONANCE IN MEDICINE//JOURNAL OF BIOMOLECULAR NMR | 54843 |
| 866 | 2 | JOURNAL OF BIOMOLECULAR NMR//JOURNAL OF MAGNETIC RESONANCE//RESIDUAL DIPOLAR COUPLINGS | 11581 |
| 19037 | 1 | BIOMAG BANK//PROT DATA BANK EUROPE//CHEMICAL SHIFT | 525 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | BIOMAG BANK | address | 759975 | 3% | 93% | 14 |
| 2 | PROT DATA BANK EUROPE | address | 518156 | 3% | 64% | 14 |
| 3 | CHEMICAL SHIFT | authKW | 371311 | 16% | 7% | 86 |
| 4 | JOURNAL OF BIOMOLECULAR NMR | journal | 369692 | 25% | 5% | 130 |
| 5 | PDBJ | address | 348969 | 1% | 100% | 6 |
| 6 | BMRB | authKW | 316654 | 1% | 78% | 7 |
| 7 | RCSB PROT DATA BANK | address | 232638 | 2% | 50% | 8 |
| 8 | PROTON CHEMICAL SHIFT | authKW | 219218 | 1% | 54% | 7 |
| 9 | PROTEIN STRUCTURE VALIDATION | authKW | 209377 | 1% | 60% | 6 |
| 10 | NOE ASSIGNMENT | authKW | 181751 | 1% | 63% | 5 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Spectroscopy | 6775 | 31% | 0% | 164 |
| 2 | Biochemistry & Molecular Biology | 2690 | 59% | 0% | 308 |
| 3 | Biophysics | 861 | 15% | 0% | 81 |
| 4 | Physics, Atomic, Molecular & Chemical | 333 | 11% | 0% | 58 |
| 5 | Biochemical Research Methods | 312 | 9% | 0% | 47 |
| 6 | Chemistry, Multidisciplinary | 235 | 16% | 0% | 85 |
| 7 | Mathematical & Computational Biology | 118 | 3% | 0% | 16 |
| 8 | Chemistry, Physical | 86 | 12% | 0% | 61 |
| 9 | Crystallography | 19 | 2% | 0% | 12 |
| 10 | Cell Biology | 8 | 4% | 0% | 22 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | BIOMAG BANK | 759975 | 3% | 93% | 14 |
| 2 | PROT DATA BANK EUROPE | 518156 | 3% | 64% | 14 |
| 3 | PDBJ | 348969 | 1% | 100% | 6 |
| 4 | RCSB PROT DATA BANK | 232638 | 2% | 50% | 8 |
| 5 | BIOMOL NMR MUNICH INTEGRATED PROT SCI | 174484 | 1% | 100% | 3 |
| 6 | PDBE | 174484 | 1% | 100% | 3 |
| 7 | PROT DATA BANK EUROPE PDBE | 174484 | 1% | 100% | 3 |
| 8 | RCSB PDB | 161555 | 1% | 56% | 5 |
| 9 | MATEMAT LICADA SAN LUIS | 145379 | 3% | 17% | 15 |
| 10 | INTEGRAT PROTE | 124984 | 2% | 18% | 12 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | JOURNAL OF BIOMOLECULAR NMR | 369692 | 25% | 5% | 130 |
| 2 | PROTEINS-STRUCTURE FUNCTION AND BIOINFORMATICS | 7637 | 6% | 0% | 29 |
| 3 | PROGRESS IN NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY | 7628 | 2% | 2% | 8 |
| 4 | JOURNAL OF MAGNETIC RESONANCE SERIES B | 3466 | 1% | 1% | 6 |
| 5 | JOURNAL OF COMPUTATIONAL CHEMISTRY | 2116 | 3% | 0% | 15 |
| 6 | PROTEIN SCIENCE | 1547 | 2% | 0% | 13 |
| 7 | JOURNAL OF MAGNETIC RESONANCE | 1436 | 3% | 0% | 15 |
| 8 | JOURNAL OF THE AMERICAN CHEMICAL SOCIETY | 1340 | 9% | 0% | 47 |
| 9 | ACTA CRYSTALLOGRAPHICA SECTION D-STRUCTURAL BIOLOGY | 1153 | 1% | 0% | 5 |
| 10 | NUCLEIC ACIDS RESEARCH | 959 | 5% | 0% | 26 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | CHEMICAL SHIFT | 371311 | 16% | 7% | 86 | Search CHEMICAL+SHIFT | Search CHEMICAL+SHIFT |
| 2 | BMRB | 316654 | 1% | 78% | 7 | Search BMRB | Search BMRB |
| 3 | PROTON CHEMICAL SHIFT | 219218 | 1% | 54% | 7 | Search PROTON+CHEMICAL+SHIFT | Search PROTON+CHEMICAL+SHIFT |
| 4 | PROTEIN STRUCTURE VALIDATION | 209377 | 1% | 60% | 6 | Search PROTEIN+STRUCTURE+VALIDATION | Search PROTEIN+STRUCTURE+VALIDATION |
| 5 | NOE ASSIGNMENT | 181751 | 1% | 63% | 5 | Search NOE+ASSIGNMENT | Search NOE+ASSIGNMENT |
| 6 | PROTEIN CHEMICAL SHIFT | 174484 | 1% | 100% | 3 | Search PROTEIN+CHEMICAL+SHIFT | Search PROTEIN+CHEMICAL+SHIFT |
| 7 | STRUCTURE VALIDATION | 167487 | 2% | 24% | 12 | Search STRUCTURE+VALIDATION | Search STRUCTURE+VALIDATION |
| 8 | CS ROSETTA | 161056 | 1% | 46% | 6 | Search CS+ROSETTA | Search CS+ROSETTA |
| 9 | CHEMICAL SHIFT PREDICTION | 153042 | 2% | 26% | 10 | Search CHEMICAL+SHIFT+PREDICTION | Search CHEMICAL+SHIFT+PREDICTION |
| 10 | BIOMAGRESBANK BMRB | 116323 | 0% | 100% | 2 | Search BIOMAGRESBANK+BMRB | Search BIOMAGRESBANK+BMRB |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | WISHART, DS , (2011) INTERPRETING PROTEIN CHEMICAL SHIFT DATA.PROGRESS IN NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY. VOL. 58. ISSUE 1-2. P. 62 -87 | 105 | 55% | 69 |
| 2 | HAN, B , LIU, YF , GINZINGER, SW , WISHART, DS , (2011) SHIFTX2: SIGNIFICANTLY IMPROVED PROTEIN CHEMICAL SHIFT PREDICTION.JOURNAL OF BIOMOLECULAR NMR. VOL. 50. ISSUE 1. P. 43 -57 | 33 | 77% | 168 |
| 3 | VUISTER, GW , FOGH, RH , HENDRICKX, PMS , DORELEIJERS, JF , GUTMANAS, A , (2014) AN OVERVIEW OF TOOLS FOR THE VALIDATION OF PROTEIN NMR STRUCTURES.JOURNAL OF BIOMOLECULAR NMR. VOL. 58. ISSUE 4. P. 259-285 | 59 | 53% | 5 |
| 4 | SHEN, Y , BAX, A , (2013) PROTEIN BACKBONE AND SIDECHAIN TORSION ANGLES PREDICTED FROM NMR CHEMICAL SHIFTS USING ARTIFICIAL NEURAL NETWORKS.JOURNAL OF BIOMOLECULAR NMR. VOL. 56. ISSUE 3. P. 227-241 | 30 | 63% | 136 |
| 5 | ZHU, T , ZHANG, JZH , HE, X , (2014) CORRECTION OF ERRONEOUSLY PACKED PROTEIN'S SIDE CHAINS IN THE NMR STRUCTURE BASED ON AB INITIO CHEMICAL SHIFT CALCULATIONS.PHYSICAL CHEMISTRY CHEMICAL PHYSICS. VOL. 16. ISSUE 34. P. 18163 -18169 | 38 | 72% | 1 |
| 6 | VRANKEN, WF , (2014) NMR STRUCTURE VALIDATION IN RELATION TO DYNAMICS AND STRUCTURE DETERMINATION.PROGRESS IN NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY. VOL. 82. ISSUE . P. 27 -38 | 47 | 52% | 1 |
| 7 | LEHTIVARJO, J , TUPPURAINEN, K , HASSINEN, T , LAATIKAINEN, R , PERAKYLA, M , (2012) COMBINING NMR ENSEMBLES AND MOLECULAR DYNAMICS SIMULATIONS PROVIDES MORE REALISTIC MODELS OF PROTEIN STRUCTURES IN SOLUTION AND LEADS TO BETTER CHEMICAL SHIFT PREDICTION.JOURNAL OF BIOMOLECULAR NMR. VOL. 52. ISSUE 3. P. 257-267 | 26 | 87% | 17 |
| 8 | ROSATO, A , TEJERO, R , MONTELIONE, GT , (2013) QUALITY ASSESSMENT OF PROTEIN NMR STRUCTURES.CURRENT OPINION IN STRUCTURAL BIOLOGY. VOL. 23. ISSUE 5. P. 715-724 | 39 | 57% | 5 |
| 9 | LI, DW , BRUSCHWEILER, R , (2015) PPM_ONE: A STATIC PROTEIN STRUCTURE BASED CHEMICAL SHIFT PREDICTOR.JOURNAL OF BIOMOLECULAR NMR. VOL. 62. ISSUE 3. P. 403 -409 | 28 | 74% | 2 |
| 10 | SHEN, Y , BAX, A , (2007) PROTEIN BACKBONE CHEMICAL SHIFTS PREDICTED FROM SEARCHING A DATABASE FOR TORSION ANGLE AND SEQUENCE HOMOLOGY.JOURNAL OF BIOMOLECULAR NMR. VOL. 38. ISSUE 4. P. 289 -302 | 31 | 62% | 172 |
Classes with closest relation at Level 1 |