Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
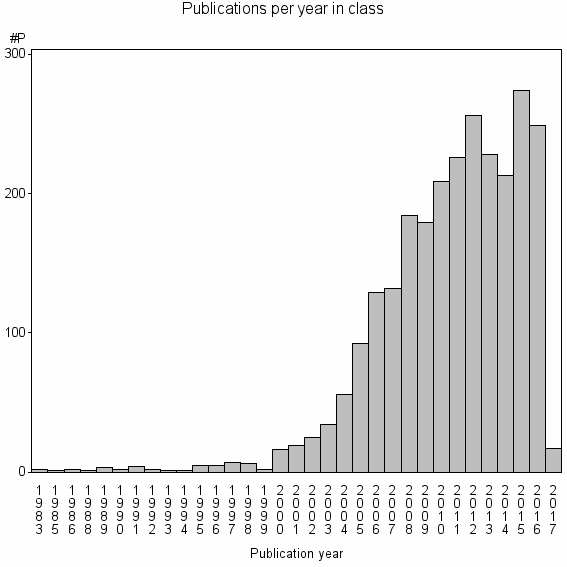
| 1585 | 2582 | 42.3 | 77% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 90 | 3 | MATHEMATICAL & COMPUTATIONAL BIOLOGY//BIOINFORMATICS//BMC BIOINFORMATICS | 77178 |
| 548 | 2 | BMC SYSTEMS BIOLOGY//MATHEMATICAL & COMPUTATIONAL BIOLOGY//BOOLEAN NETWORKS | 14655 |
| 1585 | 1 | CHEMICAL MASTER EQUATION//STOCHASTIC GENE EXPRESSION//TAU LEAPING | 2582 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | CHEMICAL MASTER EQUATION | authKW | 571386 | 2% | 79% | 61 |
| 2 | STOCHASTIC GENE EXPRESSION | authKW | 310121 | 2% | 66% | 40 |
| 3 | TAU LEAPING | authKW | 287328 | 1% | 90% | 27 |
| 4 | STOCHASTIC SIMULATION ALGORITHM | authKW | 263202 | 1% | 70% | 32 |
| 5 | LINEAR NOISE APPROXIMATION | authKW | 248825 | 1% | 96% | 22 |
| 6 | GENE EXPRESSION NOISE | authKW | 184061 | 1% | 65% | 24 |
| 7 | BIOSYST DYNAM | address | 163499 | 1% | 63% | 22 |
| 8 | STOCHASTIC CHEMICAL KINETICS | authKW | 157646 | 1% | 67% | 20 |
| 9 | INTRINSIC NOISE | authKW | 138980 | 1% | 51% | 23 |
| 10 | COMPUTAT SYST BIOL GRP | address | 71870 | 1% | 20% | 31 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Mathematical & Computational Biology | 29167 | 20% | 1% | 507 |
| 2 | Physics, Mathematical | 3977 | 12% | 0% | 297 |
| 3 | Biology | 3189 | 10% | 0% | 271 |
| 4 | Biochemical Research Methods | 2431 | 11% | 0% | 284 |
| 5 | Physics, Atomic, Molecular & Chemical | 1544 | 11% | 0% | 278 |
| 6 | Biophysics | 1488 | 10% | 0% | 249 |
| 7 | Biochemistry & Molecular Biology | 1315 | 22% | 0% | 575 |
| 8 | Physics, Fluids & Plasmas | 1307 | 6% | 0% | 155 |
| 9 | Cell Biology | 938 | 12% | 0% | 299 |
| 10 | Multidisciplinary Sciences | 639 | 3% | 0% | 75 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | BIOSYST DYNAM | 163499 | 1% | 63% | 22 |
| 2 | COMPUTAT SYST BIOL GRP | 71870 | 1% | 20% | 31 |
| 3 | CHEM PHYS PL MATH | 64447 | 1% | 42% | 13 |
| 4 | INTEGRAT SYST BIOL UNIT | 54048 | 0% | 57% | 8 |
| 5 | BIOMED ENGNMATH SCI | 35473 | 0% | 100% | 3 |
| 6 | SYNTHSYS EDINBURGH | 32841 | 0% | 56% | 5 |
| 7 | BIOCOMPLEX INFORMAT | 27700 | 1% | 11% | 21 |
| 8 | ADV COMPUTAT MODELLING | 27353 | 1% | 18% | 13 |
| 9 | THEORET BIOL PHYS | 27311 | 2% | 5% | 45 |
| 10 | GENET PHYSIOL MOL CELLULAIRE CGPHIMC | 27024 | 0% | 57% | 4 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | PHYSICAL BIOLOGY | 58305 | 2% | 8% | 64 |
| 2 | IET SYSTEMS BIOLOGY | 27871 | 1% | 8% | 28 |
| 3 | BMC SYSTEMS BIOLOGY | 24718 | 2% | 4% | 58 |
| 4 | MOLECULAR SYSTEMS BIOLOGY | 20202 | 1% | 5% | 36 |
| 5 | PLOS COMPUTATIONAL BIOLOGY | 17930 | 3% | 2% | 84 |
| 6 | MULTISCALE MODELING & SIMULATION | 6552 | 1% | 3% | 21 |
| 7 | JOURNAL OF CHEMICAL PHYSICS | 6505 | 9% | 0% | 222 |
| 8 | BIOPHYSICAL JOURNAL | 5692 | 4% | 1% | 98 |
| 9 | JOURNAL OF THE ROYAL SOCIETY INTERFACE | 5483 | 1% | 1% | 34 |
| 10 | JOURNAL OF THEORETICAL BIOLOGY | 5413 | 3% | 1% | 71 |
Author Key Words |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | GOUTSIAS, J , JENKINSON, G , (2013) MARKOVIAN DYNAMICS ON COMPLEX REACTION NETWORKS.PHYSICS REPORTS-REVIEW SECTION OF PHYSICS LETTERS. VOL. 529. ISSUE 2. P. 199 -264 | 159 | 60% | 30 |
| 2 | RAJ, A , VAN OUDENAARDEN, A , (2008) NATURE, NURTURE, OR CHANCE: STOCHASTIC GENE EXPRESSION AND ITS CONSEQUENCES.CELL. VOL. 135. ISSUE 2. P. 216-226 | 65 | 66% | 778 |
| 3 | GILLESPIE, DT , HELLANDER, A , PETZOLD, LR , (2013) PERSPECTIVE: STOCHASTIC ALGORITHMS FOR CHEMICAL KINETICS.JOURNAL OF CHEMICAL PHYSICS. VOL. 138. ISSUE 17. P. - | 62 | 93% | 63 |
| 4 | PAHLE, J , (2009) BIOCHEMICAL SIMULATIONS: STOCHASTIC, APPROXIMATE STOCHASTIC AND HYBRID APPROACHES.BRIEFINGS IN BIOINFORMATICS. VOL. 10. ISSUE 1. P. 53 -64 | 77 | 84% | 72 |
| 5 | KAERN, M , ELSTON, TC , BLAKE, WJ , COLLINS, JJ , (2005) STOCHASTICITY IN GENE EXPRESSION: FROM THEORIES TO PHENOTYPES.NATURE REVIEWS GENETICS. VOL. 6. ISSUE 6. P. 451 -464 | 65 | 53% | 1103 |
| 6 | SOLTANI, M , VARGAS-GARCIA, CA , ANTUNES, D , SINGH, A , (2016) INTERCELLULAR VARIABILITY IN PROTEIN LEVELS FROM STOCHASTIC EXPRESSION AND NOISY CELL CYCLE PROCESSES.PLOS COMPUTATIONAL BIOLOGY. VOL. 12. ISSUE 8. P. - | 75 | 76% | 0 |
| 7 | RIECKH, G , TKACIK, G , (2014) NOISE AND INFORMATION TRANSMISSION IN PROMOTERS WITH MULTIPLE INTERNAL STATES.BIOPHYSICAL JOURNAL. VOL. 106. ISSUE 5. P. 1194-1204 | 66 | 75% | 12 |
| 8 | SANCHEZ-OSORIO, I , RAMOS, F , MAYORGA, P , DANTAN, E , (2014) FOUNDATIONS FOR MODELING THE DYNAMICS OF GENE REGULATORY NETWORKS: A MULTILEVEL-PERSPECTIVE REVIEW.JOURNAL OF BIOINFORMATICS AND COMPUTATIONAL BIOLOGY. VOL. 12. ISSUE 1. P. - | 132 | 41% | 1 |
| 9 | LIU, J , FRANCOIS, JM , CAPP, JP , (2016) USE OF NOISE IN GENE EXPRESSION AS AN EXPERIMENTAL PARAMETER TO TEST PHENOTYPIC EFFECTS.YEAST. VOL. 33. ISSUE 6. P. 209 -216 | 56 | 84% | 1 |
| 10 | SINGH, A , SOLTANI, M , (2013) QUANTIFYING INTRINSIC AND EXTRINSIC VARIABILITY IN STOCHASTIC GENE EXPRESSION MODELS.PLOS ONE. VOL. 8. ISSUE 12. P. - | 54 | 96% | 5 |
Classes with closest relation at Level 1 |