Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
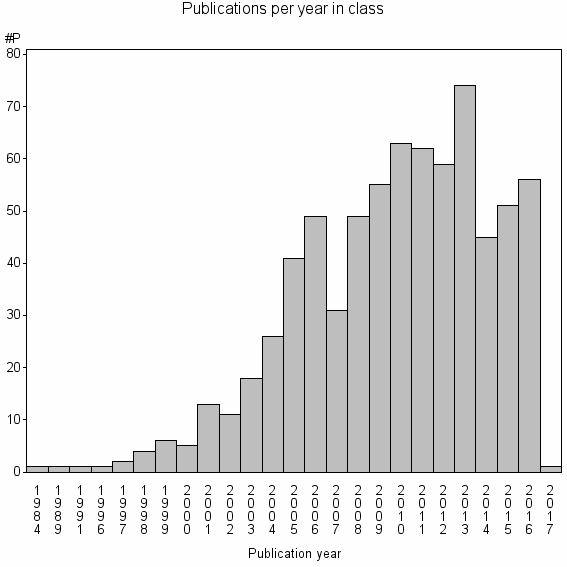
| 15220 | 725 | 36.3 | 69% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 90 | 3 | MATHEMATICAL & COMPUTATIONAL BIOLOGY//BIOINFORMATICS//BMC BIOINFORMATICS | 77178 |
| 548 | 2 | BMC SYSTEMS BIOLOGY//MATHEMATICAL & COMPUTATIONAL BIOLOGY//BOOLEAN NETWORKS | 14655 |
| 15220 | 1 | CELLML//SBML//AUCKLAND BIOENGN | 725 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | CELLML | authKW | 507142 | 2% | 71% | 17 |
| 2 | SBML | authKW | 440169 | 2% | 58% | 18 |
| 3 | AUCKLAND BIOENGN | address | 195883 | 7% | 9% | 49 |
| 4 | BIOINFORMAT TUEBINGEN ZBIT | address | 183765 | 2% | 36% | 12 |
| 5 | BIOPAX | authKW | 175483 | 1% | 83% | 5 |
| 6 | SBGN | authKW | 134771 | 1% | 80% | 4 |
| 7 | SCI DATABASES VISUALIZAT GRP | address | 134771 | 1% | 80% | 4 |
| 8 | BIOMIMET MAT ENGN BIOL TISSUES | address | 126349 | 0% | 100% | 3 |
| 9 | FIELDML | authKW | 126349 | 0% | 100% | 3 |
| 10 | PHYSIOME PROJECT | authKW | 116624 | 1% | 46% | 6 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Mathematical & Computational Biology | 42435 | 44% | 0% | 321 |
| 2 | Biochemical Research Methods | 7900 | 35% | 0% | 253 |
| 3 | Biotechnology & Applied Microbiology | 3809 | 31% | 0% | 226 |
| 4 | Computer Science, Theory & Methods | 298 | 6% | 0% | 45 |
| 5 | Computer Science, Interdisciplinary Applications | 177 | 5% | 0% | 35 |
| 6 | Biochemistry & Molecular Biology | 116 | 15% | 0% | 108 |
| 7 | Computer Science, Artificial Intelligence | 113 | 4% | 0% | 28 |
| 8 | Biology | 108 | 4% | 0% | 30 |
| 9 | Multidisciplinary Sciences | 97 | 2% | 0% | 16 |
| 10 | Engineering, Biomedical | 96 | 3% | 0% | 25 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | AUCKLAND BIOENGN | 195883 | 7% | 9% | 49 |
| 2 | BIOINFORMAT TUEBINGEN ZBIT | 183765 | 2% | 36% | 12 |
| 3 | SCI DATABASES VISUALIZAT GRP | 134771 | 1% | 80% | 4 |
| 4 | BIOMIMET MAT ENGN BIOL TISSUES | 126349 | 0% | 100% | 3 |
| 5 | BIOINFORMAT BIGCAT | 87846 | 2% | 19% | 11 |
| 6 | INTEGRAT GENET CIGENE IMT | 84233 | 0% | 100% | 2 |
| 7 | INTELLIGENT INFORMAT INFRASTRUCT | 84233 | 0% | 100% | 2 |
| 8 | GRP CLIN GENOM NETWORKS | 63172 | 0% | 50% | 3 |
| 9 | FISIOCOMP | 56154 | 0% | 67% | 2 |
| 10 | INTEGRATED OPEN SYST UNIT | 56154 | 0% | 67% | 2 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | BIOINFORMATICS | 71113 | 18% | 1% | 134 |
| 2 | BMC SYSTEMS BIOLOGY | 68281 | 7% | 3% | 51 |
| 3 | BMC BIOINFORMATICS | 13429 | 7% | 1% | 49 |
| 4 | IET SYSTEMS BIOLOGY | 10258 | 1% | 3% | 9 |
| 5 | PROGRESS IN BIOPHYSICS & MOLECULAR BIOLOGY | 8104 | 2% | 1% | 15 |
| 6 | JOURNAL OF BIOMEDICAL SEMANTICS | 6254 | 1% | 2% | 6 |
| 7 | BRIEFINGS IN BIOINFORMATICS | 4891 | 1% | 1% | 9 |
| 8 | WILEY INTERDISCIPLINARY REVIEWS-SYSTEMS BIOLOGY AND MEDICINE | 3409 | 1% | 2% | 5 |
| 9 | MOLECULAR SYSTEMS BIOLOGY | 2716 | 1% | 1% | 7 |
| 10 | DATABASE-THE JOURNAL OF BIOLOGICAL DATABASES AND CURATION | 2317 | 1% | 1% | 6 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | CELLML | 507142 | 2% | 71% | 17 | Search CELLML | Search CELLML |
| 2 | SBML | 440169 | 2% | 58% | 18 | Search SBML | Search SBML |
| 3 | BIOPAX | 175483 | 1% | 83% | 5 | Search BIOPAX | Search BIOPAX |
| 4 | SBGN | 134771 | 1% | 80% | 4 | Search SBGN | Search SBGN |
| 5 | FIELDML | 126349 | 0% | 100% | 3 | Search FIELDML | Search FIELDML |
| 6 | PHYSIOME PROJECT | 116624 | 1% | 46% | 6 | Search PHYSIOME+PROJECT | Search PHYSIOME+PROJECT |
| 7 | VIRTUAL PHYSIOLOGICAL HUMAN | 112300 | 1% | 33% | 8 | Search VIRTUAL+PHYSIOLOGICAL+HUMAN | Search VIRTUAL+PHYSIOLOGICAL+HUMAN |
| 8 | PHYSIOME | 110818 | 1% | 26% | 10 | Search PHYSIOME | Search PHYSIOME |
| 9 | RULE BASED MODELING | 89716 | 1% | 30% | 7 | Search RULE+BASED+MODELING | Search RULE+BASED+MODELING |
| 10 | BIOCHEMICAL SIMULATIONS | 84233 | 0% | 100% | 2 | Search BIOCHEMICAL+SIMULATIONS | Search BIOCHEMICAL+SIMULATIONS |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | WALTEMATH, D , BERGMANN, FT , CHAOUIYA, C , CZAUDERNA, T , GLEESON, P , GOBLE, C , GOLEBIEWSKI, M , HUCKA, M , JUTY, N , KREBS, O , ET AL (2014) MEETING REPORT FROM THE FOURTH MEETING OF THE COMPUTATIONAL MODELING IN BIOLOGY NETWORK (COMBINE).STANDARDS IN GENOMIC SCIENCES. VOL. 9. ISSUE 3. P. - | 30 | 64% | 0 |
| 2 | CHYLEK, LA , HARRIS, LA , FAEDER, JR , HLAVACEK, WS , (2015) MODELING FOR (PHYSICAL) BIOLOGISTS: AN INTRODUCTION TO THE RULE-BASED APPROACH.PHYSICAL BIOLOGY. VOL. 12. ISSUE 4. P. - | 43 | 39% | 3 |
| 3 | COSKUN, SA , CICEK, AE , LAI, N , DASH, RK , OZSOYOGLU, ZM , OZSOYOGLU, G , (2013) AN ONLINE MODEL COMPOSITION TOOL FOR SYSTEM BIOLOGY MODELS.BMC SYSTEMS BIOLOGY. VOL. 7. ISSUE . P. - | 17 | 89% | 2 |
| 4 | LI, C , DONIZELLI, M , RODRIGUEZ, N , DHARURI, H , ENDLER, L , CHELLIAH, V , LI, L , HE, EU , HENRY, A , STEFAN, MI , ET AL (2010) BIOMODELS DATABASE: AN ENHANCED, CURATED AND ANNOTATED RESOURCE FOR PUBLISHED QUANTITATIVE KINETIC MODELS.BMC SYSTEMS BIOLOGY. VOL. 4. ISSUE . P. - | 16 | 67% | 207 |
| 5 | NICKERSON, D , ATALAG, K , DE BONO, B , GEIGER, J , GOBLE, C , HOLLMANN, S , LONIEN, J , MULLER, W , REGIERER, B , STANFORD, NJ , ET AL (2016) THE HUMAN PHYSIOME: HOW STANDARDS, SOFTWARE AND INNOVATIVE SERVICE INFRASTRUCTURES ARE PROVIDING THE BUILDING BLOCKS TO MAKE IT ACHIEVABLE.INTERFACE FOCUS. VOL. 6. ISSUE 2. P. - | 24 | 51% | 2 |
| 6 | CHELLIAH, V , JUTY, N , AJMERA, I , ALI, R , DUMOUSSEAU, M , GLONT, M , HUCKA, M , JALOWICKI, G , KEATING, S , KNIGHT-SCHRIJVER, V , ET AL (2015) BIOMODELS: TEN-YEAR ANNIVERSARY.NUCLEIC ACIDS RESEARCH. VOL. 43. ISSUE D1. P. D542 -D548 | 15 | 65% | 30 |
| 7 | VAN IERSEL, MP , VILLEGER, AC , CZAUDERNA, T , BOYD, SE , BERGMANN, FT , LUNA, A , DEMIR, E , SOROKIN, A , DOGRUSOZ, U , MATSUOKA, Y , ET AL (2012) SOFTWARE SUPPORT FOR SBGN MAPS: SBGN-ML AND LIBSBGN.BIOINFORMATICS. VOL. 28. ISSUE 15. P. 2016-2021 | 14 | 100% | 12 |
| 8 | MISIRLI, G , CAVALIERE, M , WAITES, W , POCOCK, M , MADSEN, C , GILFELLON, O , HONORATO-ZIMMER, R , ZULIANI, P , DANOS, V , WIPAT, A , (2016) ANNOTATION OF RULE-BASED MODELS WITH FORMAL SEMANTICS TO ENABLE CREATION, ANALYSIS, REUSE AND VISUALIZATION.BIOINFORMATICS. VOL. 32. ISSUE 6. P. 908 -917 | 20 | 63% | 1 |
| 9 | KOLCZYK, K , CONRADI, C , (2016) CHALLENGES IN HORIZONTAL MODEL INTEGRATION.BMC SYSTEMS BIOLOGY. VOL. 10. ISSUE . P. - | 18 | 75% | 0 |
| 10 | WALTEMATH, D , WOLKENHAUER, O , (2016) HOW MODELING STANDARDS, SOFTWARE, AND INITIATIVES SUPPORT REPRODUCIBILITY IN SYSTEMS BIOLOGY AND SYSTEMS MEDICINE.IEEE TRANSACTIONS ON BIOMEDICAL ENGINEERING. VOL. 63. ISSUE 10. P. 1999 -2006 | 22 | 55% | 1 |
Classes with closest relation at Level 1 |