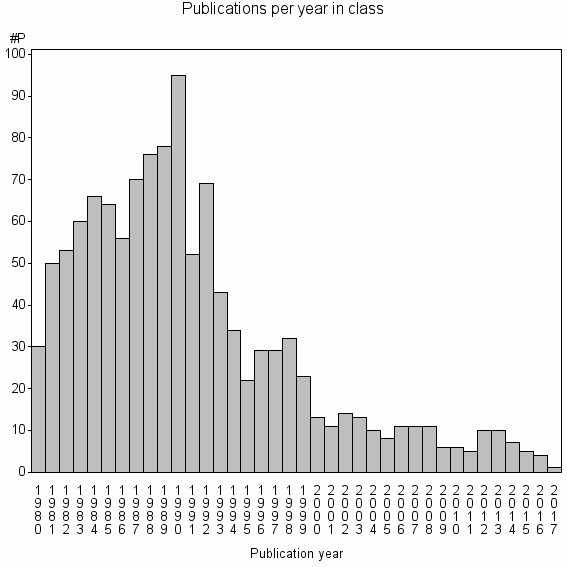
Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
| 9109 | 1187 | 39.0 | 71% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 153 | 3 | BIOCHEMISTRY & MOLECULAR BIOLOGY//RIBOSOME//RNA | 62593 |
| 156 | 2 | NUCLEAR PORE COMPLEX//PRE MRNA SPLICING//SPLICEOSOME | 22915 |
| 9109 | 1 | SNAPC//SNRNA GENES//ZNF143 | 1187 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | SNAPC | authKW | 208356 | 1% | 90% | 9 |
| 2 | SNRNA GENES | authKW | 160270 | 1% | 69% | 9 |
| 3 | ZNF143 | authKW | 96950 | 1% | 54% | 7 |
| 4 | USNRNAS | authKW | 82313 | 0% | 80% | 4 |
| 5 | PROXIMAL SEQUENCE ELEMENT | authKW | 80381 | 0% | 63% | 5 |
| 6 | INTEGRATOR COMPLEX | authKW | 68593 | 0% | 67% | 4 |
| 7 | STAF | authKW | 68593 | 0% | 67% | 4 |
| 8 | SNRNA | authKW | 64454 | 2% | 13% | 19 |
| 9 | 3 URIDYLATION | authKW | 57876 | 0% | 75% | 3 |
| 10 | BIOCHEM NC6 | address | 57876 | 0% | 75% | 3 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Biochemistry & Molecular Biology | 8177 | 67% | 0% | 797 |
| 2 | Cell Biology | 3105 | 27% | 0% | 322 |
| 3 | Rheumatology | 682 | 5% | 0% | 55 |
| 4 | Immunology | 292 | 9% | 0% | 110 |
| 5 | Genetics & Heredity | 288 | 8% | 0% | 93 |
| 6 | Biophysics | 234 | 6% | 0% | 73 |
| 7 | Developmental Biology | 106 | 2% | 0% | 27 |
| 8 | CYTOLOGY & HISTOLOGY | 76 | 0% | 0% | 5 |
| 9 | Biology | 48 | 3% | 0% | 30 |
| 10 | Biochemical Research Methods | 19 | 2% | 0% | 28 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | BIOCHEM NC6 | 57876 | 0% | 75% | 3 |
| 2 | BBRB 844 | 25723 | 0% | 100% | 1 |
| 3 | BERARD LYON | 25723 | 0% | 100% | 1 |
| 4 | BIOCHEM TRAINING PROGRAM | 25723 | 0% | 100% | 1 |
| 5 | CANC LYONCNRSINSERMU1052UMR5286 | 25723 | 0% | 100% | 1 |
| 6 | CENT IMMUNOPATHOL HEMATOL | 25723 | 0% | 100% | 1 |
| 7 | IGBMCUMR7104 | 25723 | 0% | 100% | 1 |
| 8 | IMMUNOCHIM CNRS 15 RUE DESCARTES | 25723 | 0% | 100% | 1 |
| 9 | IMTEC | 25723 | 0% | 100% | 1 |
| 10 | MEDARTHRIT IMMUNOL PROGRAM | 25723 | 0% | 100% | 1 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | NUCLEIC ACIDS RESEARCH | 17676 | 14% | 0% | 165 |
| 2 | MOLECULAR AND CELLULAR BIOLOGY | 7509 | 7% | 0% | 81 |
| 3 | MOLECULAR BIOLOGY REPORTS | 4171 | 3% | 1% | 31 |
| 4 | EMBO JOURNAL | 4161 | 4% | 0% | 53 |
| 5 | CELL | 1939 | 3% | 0% | 34 |
| 6 | GENE | 1484 | 3% | 0% | 34 |
| 7 | POSTEPY BIOCHEMII | 1332 | 0% | 3% | 2 |
| 8 | JOURNAL OF MOLECULAR BIOLOGY | 1223 | 3% | 0% | 34 |
| 9 | GENE EXPRESSION | 1221 | 0% | 1% | 4 |
| 10 | JOURNAL OF BIOLOGICAL CHEMISTRY | 1169 | 7% | 0% | 87 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | SNAPC | 208356 | 1% | 90% | 9 | Search SNAPC | Search SNAPC |
| 2 | SNRNA GENES | 160270 | 1% | 69% | 9 | Search SNRNA+GENES | Search SNRNA+GENES |
| 3 | ZNF143 | 96950 | 1% | 54% | 7 | Search ZNF143 | Search ZNF143 |
| 4 | USNRNAS | 82313 | 0% | 80% | 4 | Search USNRNAS | Search USNRNAS |
| 5 | PROXIMAL SEQUENCE ELEMENT | 80381 | 0% | 63% | 5 | Search PROXIMAL+SEQUENCE+ELEMENT | Search PROXIMAL+SEQUENCE+ELEMENT |
| 6 | INTEGRATOR COMPLEX | 68593 | 0% | 67% | 4 | Search INTEGRATOR+COMPLEX | Search INTEGRATOR+COMPLEX |
| 7 | STAF | 68593 | 0% | 67% | 4 | Search STAF | Search STAF |
| 8 | SNRNA | 64454 | 2% | 13% | 19 | Search SNRNA | Search SNRNA |
| 9 | 3 URIDYLATION | 57876 | 0% | 75% | 3 | Search 3+URIDYLATION | Search 3+URIDYLATION |
| 10 | SNRNA PROCESSING | 51446 | 0% | 100% | 2 | Search SNRNA+PROCESSING | Search SNRNA+PROCESSING |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | SOLYMOSY, F , POLLAK, T , (1993) URIDYLATE-RICH SMALL NUCLEAR RNAS (USNRNAS), THEIR GENES AND PSEUDOGENES, AND USNRNPS IN PLANTS - STRUCTURE AND FUNCTION - A COMPARATIVE APPROACH.CRITICAL REVIEWS IN PLANT SCIENCES. VOL. 12. ISSUE 4. P. 275 -369 | 219 | 46% | 36 |
| 2 | ZIEVE, GW , SAUTERER, RA , (1990) CELL BIOLOGY OF THE SNRNP PARTICLES.CRITICAL REVIEWS IN BIOCHEMISTRY AND MOLECULAR BIOLOGY. VOL. 25. ISSUE 1. P. 1-46 | 146 | 54% | 114 |
| 3 | HUNG, KH , STUMPH, WE , (2011) REGULATION OF SNRNA GENE EXPRESSION BY THE DROSOPHILA MELANOGASTER SMALL NUCLEAR RNA ACTIVATING PROTEIN COMPLEX (DMSNAPC).CRITICAL REVIEWS IN BIOCHEMISTRY AND MOLECULAR BIOLOGY. VOL. 46. ISSUE 1. P. 11-26 | 43 | 65% | 10 |
| 4 | JAWDEKAR, GW , HENRY, RW , (2008) TRANSCRIPTIONAL REGULATION OF HUMAN SMALL NUCLEAR RNA GENES.BIOCHIMICA ET BIOPHYSICA ACTA-GENE REGULATORY MECHANISMS. VOL. 1779. ISSUE 5. P. 295-305 | 69 | 41% | 31 |
| 5 | REDDY, R , (1989) COMPILATION OF SMALL NUCLEAR-RNA SEQUENCES.METHODS IN ENZYMOLOGY. VOL. 180. ISSUE . P. 521-532 | 61 | 86% | 12 |
| 6 | HERNANDEZ, G , VALAFAR, F , STUMPH, WE , (2007) INSECT SMALL NUCLEAR RNA GENE PROMOTERS EVOLVE RAPIDLY YET RETAIN CONSERVED FEATURES INVOLVED IN DETERMINING PROMOTER ACTIVITY AND RNA POLYMERASE SPECIFICITY.NUCLEIC ACIDS RESEARCH. VOL. 35. ISSUE 1. P. 21-34 | 36 | 73% | 11 |
| 7 | KIM, MK , KANG, YS , LAI, HT , BARAKAT, NH , MAGANTE, D , STUMPH, WE , (2010) IDENTIFICATION OF SNAPC SUBUNIT DOMAINS THAT INTERACT WITH SPECIFIC NUCLEOTIDE POSITIONS IN THE U1 AND U6 GENE PROMOTERS.MOLECULAR AND CELLULAR BIOLOGY. VOL. 30. ISSUE 10. P. 2411 -2423 | 27 | 87% | 7 |
| 8 | HUNG, KH , TITUS, M , CHIANG, SC , STUMPH, WE , (2009) A MAP OF DROSOPHILA MELANOGASTER SMALL NUCLEAR RNA-ACTIVATING PROTEIN COMPLEX (DMSNAPC) DOMAINS INVOLVED IN SUBUNIT ASSEMBLY AND DNA BINDING.JOURNAL OF BIOLOGICAL CHEMISTRY. VOL. 284. ISSUE 34. P. 22568 -22579 | 25 | 89% | 3 |
| 9 | LUHRMANN, R , KASTNER, B , BACH, M , (1990) STRUCTURE OF SPLICEOSOMAL SNRNPS AND THEIR ROLE IN PRE-MESSENGER-RNA SPLICING.BIOCHIMICA ET BIOPHYSICA ACTA. VOL. 1087. ISSUE 3. P. 265-292 | 71 | 41% | 319 |
| 10 | LU, ZP , MATERA, AG , (2015) DEVELOPMENTAL ANALYSIS OF SPLICEOSOMAL SNRNA LSOFORM EXPRESSION.G3-GENES GENOMES GENETICS. VOL. 5. ISSUE 1. P. 103 -110 | 30 | 61% | 0 |
Classes with closest relation at Level 1 |