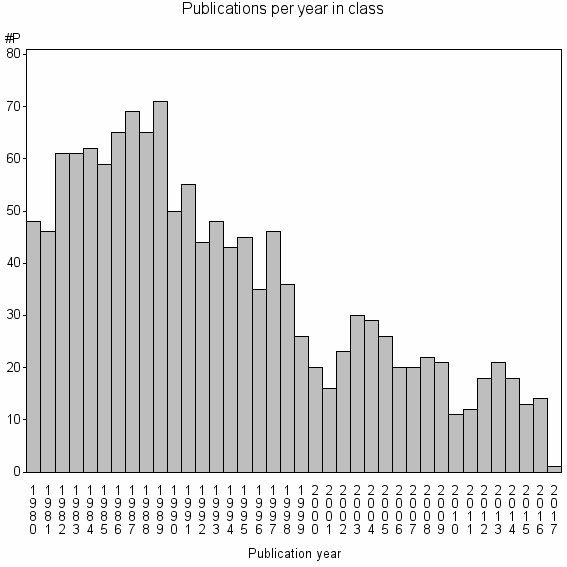
Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
| 7407 | 1370 | 40.3 | 69% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 71 | 3 | GENETICS & HEREDITY//CHROMATIN//CELL BIOLOGY | 83302 |
| 1094 | 2 | NUCLEAR MATRIX//LINKER HISTONE//HISTONE H1 | 9829 |
| 7407 | 1 | HISTONE MRNA//HISTONE GENES//HISTONE GENE EXPRESSION | 1370 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | HISTONE MRNA | authKW | 466452 | 2% | 70% | 30 |
| 2 | HISTONE GENES | authKW | 451279 | 3% | 56% | 36 |
| 3 | HISTONE GENE EXPRESSION | authKW | 383119 | 1% | 90% | 19 |
| 4 | SLBP | authKW | 328219 | 1% | 82% | 18 |
| 5 | PROGRAM MOL BIOL BIOTECHNOL | address | 319043 | 5% | 20% | 72 |
| 6 | U7 SNRNP | authKW | 269033 | 1% | 93% | 13 |
| 7 | STEM LOOP BINDING PROTEIN | authKW | 179775 | 1% | 73% | 11 |
| 8 | AD T EVOLUT | address | 136506 | 1% | 88% | 7 |
| 9 | NPAT | authKW | 134826 | 1% | 55% | 11 |
| 10 | HISTONE H1T | authKW | 129665 | 1% | 73% | 8 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Biochemistry & Molecular Biology | 9597 | 68% | 0% | 927 |
| 2 | Cell Biology | 6377 | 35% | 0% | 485 |
| 3 | Developmental Biology | 2136 | 8% | 0% | 113 |
| 4 | Genetics & Heredity | 1381 | 14% | 0% | 196 |
| 5 | Biophysics | 527 | 8% | 0% | 111 |
| 6 | Evolutionary Biology | 212 | 3% | 0% | 41 |
| 7 | Plant Sciences | 146 | 6% | 0% | 84 |
| 8 | Biology | 113 | 3% | 0% | 45 |
| 9 | Reproductive Biology | 69 | 2% | 0% | 24 |
| 10 | CYTOLOGY & HISTOLOGY | 40 | 0% | 0% | 4 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | PROGRAM MOL BIOL BIOTECHNOL | 319043 | 5% | 20% | 72 |
| 2 | AD T EVOLUT | 136506 | 1% | 88% | 7 |
| 3 | INTEGRAT PROGRAM BIOL GENOME SCI | 70951 | 1% | 29% | 11 |
| 4 | ZOOL ABT ENTWICKLUNGSBIOL | 44574 | 0% | 100% | 2 |
| 5 | HEMATOL BEIJING CHILDRENS HOSP | 22287 | 0% | 100% | 1 |
| 6 | MOL BIOL BIOCHEM MOL CELL BIOL | 22287 | 0% | 100% | 1 |
| 7 | SCI ANIM EXPTERIMENTAT | 22287 | 0% | 100% | 1 |
| 8 | STEM CELL REGENERAT MED TISSUE ENGN | 22287 | 0% | 100% | 1 |
| 9 | GRP CELLULAR SENESCENCE | 16711 | 0% | 25% | 3 |
| 10 | BIOCHEM MOL ZELLBIOL | 16626 | 1% | 7% | 11 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | MOLECULAR AND CELLULAR BIOLOGY | 12258 | 8% | 1% | 111 |
| 2 | NUCLEIC ACIDS RESEARCH | 9425 | 9% | 0% | 130 |
| 3 | JOURNAL OF CELLULAR BIOCHEMISTRY | 5467 | 3% | 1% | 47 |
| 4 | JOURNAL OF MOLECULAR EVOLUTION | 5219 | 2% | 1% | 30 |
| 5 | DEVELOPMENTAL BIOLOGY | 4843 | 4% | 0% | 55 |
| 6 | PLANT MOLECULAR BIOLOGY | 2383 | 2% | 0% | 27 |
| 7 | RNA | 2223 | 1% | 1% | 16 |
| 8 | GENE | 1967 | 3% | 0% | 42 |
| 9 | PROCEEDINGS OF THE NATIONAL ACADEMY OF SCIENCES OF THE UNITED STATES OF AMERICA-BIOLOGICAL SCIENCES | 1864 | 2% | 0% | 26 |
| 10 | BIOCHIMICA ET BIOPHYSICA ACTA-GENE STRUCTURE AND EXPRESSION | 1819 | 1% | 1% | 15 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | HISTONE MRNA | 466452 | 2% | 70% | 30 | Search HISTONE+MRNA | Search HISTONE+MRNA |
| 2 | HISTONE GENES | 451279 | 3% | 56% | 36 | Search HISTONE+GENES | Search HISTONE+GENES |
| 3 | HISTONE GENE EXPRESSION | 383119 | 1% | 90% | 19 | Search HISTONE+GENE+EXPRESSION | Search HISTONE+GENE+EXPRESSION |
| 4 | SLBP | 328219 | 1% | 82% | 18 | Search SLBP | Search SLBP |
| 5 | U7 SNRNP | 269033 | 1% | 93% | 13 | Search U7+SNRNP | Search U7+SNRNP |
| 6 | STEM LOOP BINDING PROTEIN | 179775 | 1% | 73% | 11 | Search STEM+LOOP+BINDING+PROTEIN | Search STEM+LOOP+BINDING+PROTEIN |
| 7 | NPAT | 134826 | 1% | 55% | 11 | Search NPAT | Search NPAT |
| 8 | HISTONE H1T | 129665 | 1% | 73% | 8 | Search HISTONE+H1T | Search HISTONE+H1T |
| 9 | CASP8AP2 | 114617 | 0% | 86% | 6 | Search CASP8AP2 | Search CASP8AP2 |
| 10 | HISTONE H1T GENE | 111435 | 0% | 100% | 5 | Search HISTONE+H1T+GENE | Search HISTONE+H1T+GENE |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | OSLEY, MA , (1991) THE REGULATION OF HISTONE SYNTHESIS IN THE CELL-CYCLE.ANNUAL REVIEW OF BIOCHEMISTRY. VOL. 60. ISSUE . P. 827-861 | 119 | 59% | 402 |
| 2 | MARZLUFF, WF , WAGNER, EJ , DURONIO, RJ , (2008) METABOLISM AND REGULATION OF CANONICAL HISTONE MRNAS: LIFE WITHOUT A POLY(A) TAIL.NATURE REVIEWS GENETICS. VOL. 9. ISSUE 11. P. 843-854 | 62 | 63% | 229 |
| 3 | DOMINSKI, Z , MARZLUFF, WF , (1999) FORMATION OF THE 3 ' END OF HISTONE MRNA.GENE. VOL. 239. ISSUE 1. P. 1-14 | 75 | 80% | 125 |
| 4 | DOMINSKI, Z , MARZLUFF, WF , (2007) FORMATION OF THE 3 ' END OF HISTONE MRNA: GETTING CLOSER TO THE END.GENE. VOL. 396. ISSUE 2. P. 373-390 | 87 | 51% | 66 |
| 5 | MESHI, T , TAOKA, K , IWABUCHI, M , (2000) REGULATION OF HISTONE GENE EXPRESSION DURING THE CELL CYCLE.PLANT MOLECULAR BIOLOGY. VOL. 43. ISSUE 5-6. P. 643 -657 | 71 | 74% | 37 |
| 6 | LIU, LJ , XIE, RL , HUSSAIN, S , LIAN, JB , RIVERA-PEREZ, J , JONES, SN , STEIN, JL , STEIN, GS , VAN WIJNEN, AJ , (2011) FUNCTIONAL COUPLING OF TRANSCRIPTION FACTOR HINF-P AND HISTONE H4 GENE EXPRESSION DURING PRE- AND POST-NATAL MOUSE DEVELOPMENT.GENE. VOL. 483. ISSUE 1-2. P. 1-10 | 50 | 77% | 2 |
| 7 | LYONS, SM , CUNNINGHAM, CH , WELCH, JD , GROH, B , GUO, AY , WEI, B , WHITFIELD, ML , XIONG, Y , MARZLUFF, WF , (2016) A SUBSET OF REPLICATION-DEPENDENT HISTONE MRNAS ARE EXPRESSED AS POLYADENYLATED RNAS IN TERMINALLY DIFFERENTIATED TISSUES.NUCLEIC ACIDS RESEARCH. VOL. 44. ISSUE 19. P. 9190 -9205 | 49 | 64% | 1 |
| 8 | DOMINSKI, Z , YANG, XC , RASKA, CS , SANTIAGO, C , BORCHERS, CH , DURONIO, RJ , MARZLUFF, WF , (2002) 3 ' END PROCESSING OF DROSOPHILA MELANOGASTER HISTONE PRE-MRNAS: REQUIREMENT FOR PHOSPHORYLATED DROSOPHILA STEM-LOOP BINDING PROTEIN AND COEVOLUTION OF THE HISTONE PRE-MRNA PROCESSING SYSTEM.MOLECULAR AND CELLULAR BIOLOGY. VOL. 22. ISSUE 18. P. 6648-6660 | 54 | 84% | 23 |
| 9 | ROMEO, V , SCHUMPERLI, D , (2016) CYCLING IN THE NUCLEUS: REGULATION OF RNA 30 PROCESSING AND NUCLEAR ORGANIZATION OF REPLICATION-DEPENDENT HISTONE GENES.CURRENT OPINION IN CELL BIOLOGY. VOL. 40. ISSUE . P. 23 -31 | 40 | 63% | 4 |
| 10 | WELLS, D , MCBRIDE, C , (1989) A COMPREHENSIVE COMPILATION AND ALIGNMENT OF HISTONES AND HISTONE GENES.NUCLEIC ACIDS RESEARCH. VOL. 17. ISSUE . P. R311-R346 | 81 | 78% | 97 |
Classes with closest relation at Level 1 |