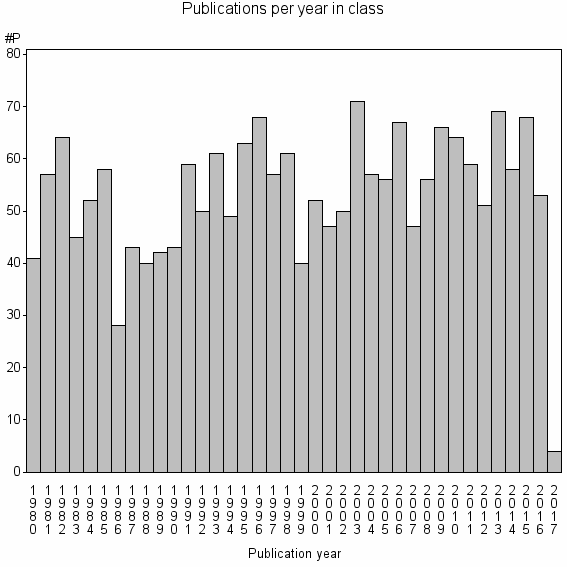
Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
| 3302 | 2016 | 44.3 | 75% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 7 | 4 | INFECTIOUS DISEASES//MICROBIOLOGY//VIROLOGY | 1353914 |
| 33 | 3 | JOURNAL OF BACTERIOLOGY//MICROBIOLOGY//MOLECULAR MICROBIOLOGY | 107033 |
| 371 | 2 | RNA POLYMERASE//HFQ//JOURNAL OF BACTERIOLOGY | 17271 |
| 3302 | 1 | RNA POLYMERASE//TRANSCRIPTION TERMINATION//NUSG | 2016 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | RNA POLYMERASE | authKW | 401792 | 9% | 15% | 177 |
| 2 | TRANSCRIPTION TERMINATION | authKW | 343798 | 4% | 32% | 71 |
| 3 | NUSG | authKW | 332429 | 1% | 73% | 30 |
| 4 | ANTITERMINATION | authKW | 232913 | 2% | 42% | 37 |
| 5 | NUSA | authKW | 225348 | 1% | 60% | 25 |
| 6 | RHO FACTOR | authKW | 223034 | 1% | 82% | 18 |
| 7 | RHO DEPENDENT TERMINATION | authKW | 136303 | 0% | 100% | 9 |
| 8 | TRANSCRIPT BIOL | address | 121869 | 1% | 62% | 13 |
| 9 | TRANSCRIPTIONAL PAUSING | authKW | 118722 | 1% | 56% | 14 |
| 10 | NUS FACTORS | authKW | 106013 | 0% | 100% | 7 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Biochemistry & Molecular Biology | 17079 | 74% | 0% | 1489 |
| 2 | Cell Biology | 1860 | 17% | 0% | 344 |
| 3 | Microbiology | 1435 | 12% | 0% | 249 |
| 4 | Genetics & Heredity | 830 | 10% | 0% | 196 |
| 5 | Biophysics | 536 | 7% | 0% | 140 |
| 6 | Biology | 159 | 3% | 0% | 65 |
| 7 | Biochemical Research Methods | 127 | 4% | 0% | 73 |
| 8 | Developmental Biology | 25 | 1% | 0% | 22 |
| 9 | Mathematical & Computational Biology | 19 | 1% | 0% | 18 |
| 10 | Virology | 5 | 1% | 0% | 19 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | TRANSCRIPT BIOL | 121869 | 1% | 62% | 13 |
| 2 | ITP SCI BIOL CHIM VIVANT | 75724 | 0% | 100% | 5 |
| 3 | FOR UNGSZENTRUM BIOMAKROMOL | 69226 | 0% | 57% | 8 |
| 4 | SECT MICROBIAL GENET | 67458 | 0% | 64% | 7 |
| 5 | GENE REGULAT CHROMOSOME BIOL | 42024 | 1% | 11% | 25 |
| 6 | LEHRSTUHL BIOPOLYMERE | 32427 | 1% | 14% | 15 |
| 7 | COMPARAT PROTEOM | 30289 | 0% | 100% | 2 |
| 8 | ECOLE DOCTORALE SANTE SCI BIOL CHIM VIVANT ED 5 | 30289 | 0% | 100% | 2 |
| 9 | EDWARD A DOISEY BIOCHEM | 30289 | 0% | 100% | 2 |
| 10 | FOR UNGSZENTRUM BIO MAKROMOL | 30289 | 0% | 100% | 2 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | JOURNAL OF MOLECULAR BIOLOGY | 22856 | 9% | 1% | 188 |
| 2 | JOURNAL OF BIOLOGICAL CHEMISTRY | 7160 | 13% | 0% | 272 |
| 3 | CELL | 5905 | 4% | 1% | 77 |
| 4 | JOURNAL OF BACTERIOLOGY | 5895 | 6% | 0% | 115 |
| 5 | MOLECULAR CELL | 5805 | 2% | 1% | 46 |
| 6 | NUCLEIC ACIDS RESEARCH | 4328 | 5% | 0% | 108 |
| 7 | MOLECULAR AND GENERAL GENETICS | 3806 | 2% | 1% | 35 |
| 8 | PROCEEDINGS OF THE NATIONAL ACADEMY OF SCIENCES OF THE UNITED STATES OF AMERICA | 2914 | 7% | 0% | 139 |
| 9 | BIOCHIMICA ET BIOPHYSICA ACTA-GENE REGULATORY MECHANISMS | 2059 | 1% | 1% | 12 |
| 10 | MOLECULAR MICROBIOLOGY | 1975 | 2% | 0% | 40 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | RNA POLYMERASE | 401792 | 9% | 15% | 177 | Search RNA+POLYMERASE | Search RNA+POLYMERASE |
| 2 | TRANSCRIPTION TERMINATION | 343798 | 4% | 32% | 71 | Search TRANSCRIPTION+TERMINATION | Search TRANSCRIPTION+TERMINATION |
| 3 | NUSG | 332429 | 1% | 73% | 30 | Search NUSG | Search NUSG |
| 4 | ANTITERMINATION | 232913 | 2% | 42% | 37 | Search ANTITERMINATION | Search ANTITERMINATION |
| 5 | NUSA | 225348 | 1% | 60% | 25 | Search NUSA | Search NUSA |
| 6 | RHO FACTOR | 223034 | 1% | 82% | 18 | Search RHO+FACTOR | Search RHO+FACTOR |
| 7 | RHO DEPENDENT TERMINATION | 136303 | 0% | 100% | 9 | Search RHO+DEPENDENT+TERMINATION | Search RHO+DEPENDENT+TERMINATION |
| 8 | TRANSCRIPTIONAL PAUSING | 118722 | 1% | 56% | 14 | Search TRANSCRIPTIONAL+PAUSING | Search TRANSCRIPTIONAL+PAUSING |
| 9 | NUS FACTORS | 106013 | 0% | 100% | 7 | Search NUS+FACTORS | Search NUS+FACTORS |
| 10 | TRANSCRIPT ELONGATION | 102343 | 1% | 48% | 14 | Search TRANSCRIPT+ELONGATION | Search TRANSCRIPT+ELONGATION |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | RAY-SONI, A , BELLECOURT, MJ , LANDICK, R , (2016) MECHANISMS OF BACTERIAL TRANSCRIPTION TERMINATION: ALL GOOD THINGS MUST END.ANNUAL REVIEW OF BIOCHEMISTRY, VOL 85. VOL. 85. ISSUE . P. 319 -347 | 124 | 86% | 3 |
| 2 | PETERS, JM , VANGELOFF, AD , LANDICK, R , (2011) BACTERIAL TRANSCRIPTION TERMINATORS: THE RNA 3 '-END CHRONICLES.JOURNAL OF MOLECULAR BIOLOGY. VOL. 412. ISSUE 5. P. 793-813 | 106 | 88% | 89 |
| 3 | NUDLER, E , GOTTESMAN, ME , (2002) TRANSCRIPTION TERMINATION AND ANTI-TERMINATION IN E-COLI.GENES TO CELLS. VOL. 7. ISSUE 8. P. 755-768 | 127 | 92% | 115 |
| 4 | ROBERTS, JW , SHANKAR, S , FILTER, JJ , (2008) RNA POLYMERASE ELONGATION FACTORS.ANNUAL REVIEW OF MICROBIOLOGY. VOL. 62. ISSUE . P. 211-233 | 119 | 82% | 63 |
| 5 | MARTINEZ-RUCOBO, FW , CRAMER, P , (2013) STRUCTURAL BASIS OF TRANSCRIPTION ELONGATION.BIOCHIMICA ET BIOPHYSICA ACTA-GENE REGULATORY MECHANISMS. VOL. 1829. ISSUE 1. P. 9-19 | 109 | 70% | 28 |
| 6 | MA, C , YANG, X , LEWIS, PJ , (2016) BACTERIAL TRANSCRIPTION AS A TARGET FOR ANTIBACTERIAL DRUG DEVELOPMENT.MICROBIOLOGY AND MOLECULAR BIOLOGY REVIEWS. VOL. 80. ISSUE 1. P. 139 -160 | 120 | 61% | 1 |
| 7 | NUDLER, E , (2009) RNA POLYMERASE ACTIVE CENTER: THE MOLECULAR ENGINE OF TRANSCRIPTION.ANNUAL REVIEW OF BIOCHEMISTRY. VOL. 78. ISSUE . P. 335-361 | 104 | 76% | 60 |
| 8 | FISH, RN , KANE, CM , (2002) PROMOTING ELONGATION WITH TRANSCRIPT CLEAVAGE STIMULATORY FACTORS.BIOCHIMICA ET BIOPHYSICA ACTA-GENE STRUCTURE AND EXPRESSION. VOL. 1577. ISSUE 2. P. 287-307 | 131 | 70% | 144 |
| 9 | PROSHKIN, SA , MIRONOV, AS , (2011) REGULATION OF BACTERIAL TRANSCRIPTION ELONGATION.MOLECULAR BIOLOGY. VOL. 45. ISSUE 3. P. 355-374 | 104 | 74% | 0 |
| 10 | BELOGUROV, GA , ARTSIMOVITCH, I , (2015) REGULATION OF TRANSCRIPT ELONGATION.ANNUAL REVIEW OF MICROBIOLOGY, VOL 69. VOL. 69. ISSUE . P. 49 -69 | 93 | 63% | 8 |
Classes with closest relation at Level 1 |