Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
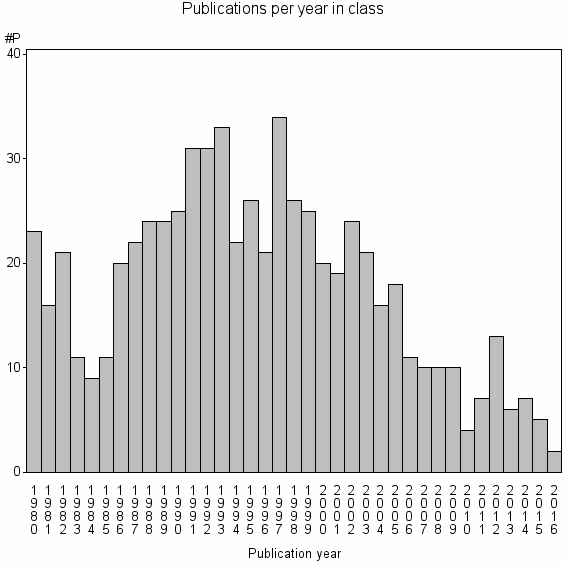
| 16354 | 658 | 31.9 | 66% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 7 | 4 | INFECTIOUS DISEASES//MICROBIOLOGY//VIROLOGY | 1353914 |
| 33 | 3 | JOURNAL OF BACTERIOLOGY//MICROBIOLOGY//MOLECULAR MICROBIOLOGY | 107033 |
| 959 | 2 | BACTERIOPHAGE//PHAGE THERAPY//ENDOLYSIN | 10780 |
| 16354 | 1 | T7 RNA POLYMERASE//T7 LYSOZYME//MORSE MOL GENET | 658 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | T7 RNA POLYMERASE | authKW | 984204 | 10% | 31% | 68 |
| 2 | T7 LYSOZYME | authKW | 330287 | 2% | 65% | 11 |
| 3 | MORSE MOL GENET | address | 304776 | 3% | 39% | 17 |
| 4 | PHAGE RNA POLYMERASE | authKW | 284229 | 1% | 88% | 7 |
| 5 | DNTP UTILIZATION | authKW | 139215 | 0% | 100% | 3 |
| 6 | K11 RNA POLYMERASE | authKW | 139215 | 0% | 100% | 3 |
| 7 | BACTERIOPHAGE T7 | authKW | 133628 | 2% | 24% | 12 |
| 8 | ABORTIVE TRANSCRIPTION | authKW | 123744 | 1% | 67% | 4 |
| 9 | BACTERIOPHAGE T7 RNA POLYMERASE | authKW | 123744 | 1% | 67% | 4 |
| 10 | GENE TANDEM EXPRESSION | authKW | 92810 | 0% | 100% | 2 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Biochemistry & Molecular Biology | 5402 | 73% | 0% | 479 |
| 2 | Virology | 486 | 7% | 0% | 46 |
| 3 | Biochemical Research Methods | 281 | 8% | 0% | 51 |
| 4 | Genetics & Heredity | 223 | 9% | 0% | 59 |
| 5 | Biotechnology & Applied Microbiology | 155 | 8% | 0% | 52 |
| 6 | Biophysics | 151 | 7% | 0% | 43 |
| 7 | Cell Biology | 87 | 8% | 0% | 52 |
| 8 | Microbiology | 65 | 6% | 0% | 37 |
| 9 | Biology | 33 | 3% | 0% | 18 |
| 10 | Crystallography | 0 | 1% | 0% | 6 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | MORSE MOL GENET | 304776 | 3% | 39% | 17 |
| 2 | BIOMINERALISAT MORPHOGENESE DIATOMEES | 46405 | 0% | 100% | 1 |
| 3 | CNRS URA 1302 | 46405 | 0% | 100% | 1 |
| 4 | INGN ANTICORPS SANTEEQUIPE MIXTE CEA BIOMER | 46405 | 0% | 100% | 1 |
| 5 | PAIK HOSP MOLEC BIOL | 46405 | 0% | 100% | 1 |
| 6 | HIGH THROUGHPUT TORY | 26514 | 0% | 29% | 2 |
| 7 | MOL BIOL 48 | 15467 | 0% | 33% | 1 |
| 8 | UMR 8183 | 15467 | 0% | 33% | 1 |
| 9 | BK21 VET SCI | 11600 | 0% | 25% | 1 |
| 10 | MOL INFORMAT PROC | 11600 | 0% | 25% | 1 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | JOURNAL OF MOLECULAR BIOLOGY | 11786 | 12% | 0% | 77 |
| 2 | MOLECULAR BIOLOGY | 3545 | 3% | 0% | 19 |
| 3 | BIOCHEMISTRY | 2578 | 8% | 0% | 55 |
| 4 | NUCLEIC ACIDS RESEARCH | 2223 | 7% | 0% | 44 |
| 5 | BIOTECHNIQUES | 1936 | 2% | 0% | 16 |
| 6 | GENE | 1711 | 4% | 0% | 27 |
| 7 | JOURNAL OF BIOLOGICAL CHEMISTRY | 1382 | 10% | 0% | 69 |
| 8 | GENE EXPRESSION | 550 | 0% | 1% | 2 |
| 9 | METHODS IN ENZYMOLOGY | 535 | 2% | 0% | 14 |
| 10 | ZEITSCHRIFT FUR ALLGEMEINE MIKROBIOLOGIE | 494 | 0% | 1% | 2 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | T7 RNA POLYMERASE | 984204 | 10% | 31% | 68 | Search T7+RNA+POLYMERASE | Search T7+RNA+POLYMERASE |
| 2 | T7 LYSOZYME | 330287 | 2% | 65% | 11 | Search T7+LYSOZYME | Search T7+LYSOZYME |
| 3 | PHAGE RNA POLYMERASE | 284229 | 1% | 88% | 7 | Search PHAGE+RNA+POLYMERASE | Search PHAGE+RNA+POLYMERASE |
| 4 | DNTP UTILIZATION | 139215 | 0% | 100% | 3 | Search DNTP+UTILIZATION | Search DNTP+UTILIZATION |
| 5 | K11 RNA POLYMERASE | 139215 | 0% | 100% | 3 | Search K11+RNA+POLYMERASE | Search K11+RNA+POLYMERASE |
| 6 | BACTERIOPHAGE T7 | 133628 | 2% | 24% | 12 | Search BACTERIOPHAGE+T7 | Search BACTERIOPHAGE+T7 |
| 7 | ABORTIVE TRANSCRIPTION | 123744 | 1% | 67% | 4 | Search ABORTIVE+TRANSCRIPTION | Search ABORTIVE+TRANSCRIPTION |
| 8 | BACTERIOPHAGE T7 RNA POLYMERASE | 123744 | 1% | 67% | 4 | Search BACTERIOPHAGE+T7+RNA+POLYMERASE | Search BACTERIOPHAGE+T7+RNA+POLYMERASE |
| 9 | GENE TANDEM EXPRESSION | 92810 | 0% | 100% | 2 | Search GENE+TANDEM+EXPRESSION | Search GENE+TANDEM+EXPRESSION |
| 10 | PHAGE K11 LYSOZYME | 92810 | 0% | 100% | 2 | Search PHAGE+K11+LYSOZYME | Search PHAGE+K11+LYSOZYME |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | IMBURGIO, D , RONG, MQ , MA, KY , MCALLISTER, WT , (2000) STUDIES OF PROMOTER RECOGNITION AND START SITE SELECTION BY T7 RNA POLYMERASE USING A COMPREHENSIVE COLLECTION OF PROMOTER VARIANTS.BIOCHEMISTRY. VOL. 39. ISSUE 34. P. 10419 -10430 | 48 | 92% | 81 |
| 2 | SOUSA, R , MUKHERJEE, S , (2003) T7 RNA POLYMERASE.PROGRESS IN NUCLEIC ACID RESEARCH AND MOLECULAR BIOLOGY, VOL 73. VOL. 73. ISSUE . P. 1 -41 | 69 | 63% | 27 |
| 3 | BANDWAR, RP , TANG, GQ , PATEL, SS , (2006) SEQUENTIAL RELEASE OF PROMOTER CONTACTS DURING TRANSCRIPTION INITIATION TO ELONGATION TRANSITION.JOURNAL OF MOLECULAR BIOLOGY. VOL. 360. ISSUE 2. P. 466 -483 | 42 | 95% | 5 |
| 4 | RUSAKOVA, EE , TUNITSKAYA, VL , KOCHETKOV, SN , (1999) T7 RNA POLYMERASE.MOLECULAR BIOLOGY. VOL. 33. ISSUE 3. P. 304 -317 | 61 | 74% | 0 |
| 5 | MARTIN, CT , ESPOSITO, EA , THEIS, K , GONG, P , (2005) STRUCTURE AND FUNCTION IN PROMOTER ESCAPE BY T7 RNA POLYMERASE.PROGRESS IN NUCLEIC ACID RESEARCH AND MOLECULAR BIOLOGY, VOL 80. VOL. 80. ISSUE . P. 323 -+ | 45 | 79% | 7 |
| 6 | MENTESANA, PE , CHIN-BOW, ST , SOUSA, R , MCALLISTER, WT , (2000) CHARACTERIZATION OF HALTED T7 RNA POLYMERASE ELONGATION COMPLEXES REVEALS MULTIPLE FACTORS THAT CONTRIBUTE TO STABILITY.JOURNAL OF MOLECULAR BIOLOGY. VOL. 302. ISSUE 5. P. 1049 -1062 | 41 | 91% | 28 |
| 7 | ZHANG, X , STUDIER, FW , (2004) MULTIPLE ROLES OF T7 RNA POLYMERASE AND T7 LYSOZYME DURING BACTERIOPHAGE T7 INFECTION.JOURNAL OF MOLECULAR BIOLOGY. VOL. 340. ISSUE 4. P. 707-730 | 40 | 80% | 28 |
| 8 | JIANG, ML , RONG, MQ , MARTIN, C , MCALLISTER, WT , (2001) INTERRUPTING THE TEMPLATE STRAND OF THE T7 PROMOTER FACILITATES TRANSLOCATION OF THE DNA DURING INITIATION, REDUCING TRANSCRIPT SLIPPAGE AND THE RELEASE OF ABORTIVE PRODUCTS.JOURNAL OF MOLECULAR BIOLOGY. VOL. 310. ISSUE 3. P. 509 -522 | 32 | 97% | 18 |
| 9 | HUANG, JB , VILLEMAIN, J , PADILLA, R , SOUSA, R , (1999) MECHANISMS BY WHICH T7 LYSOZYME SPECIFICALLY REGULATES T7 RNA POLYMERASE DURING DIFFERENT PHASES OF TRANSCRIPTION.JOURNAL OF MOLECULAR BIOLOGY. VOL. 293. ISSUE 3. P. 457-475 | 35 | 92% | 21 |
| 10 | BANDWAR, RP , MA, N , EMANUEL, SA , ANIKIN, M , VASSYLYEV, DG , PATEL, SS , MCALLISTER, WT , (2007) THE TRANSITION TO AN ELONGATION COMPLEX BY T7 RNA POLYMERASE IS A MULTISTEP PROCESS.JOURNAL OF BIOLOGICAL CHEMISTRY. VOL. 282. ISSUE 31. P. 22879-22886 | 29 | 88% | 7 |
Classes with closest relation at Level 1 |