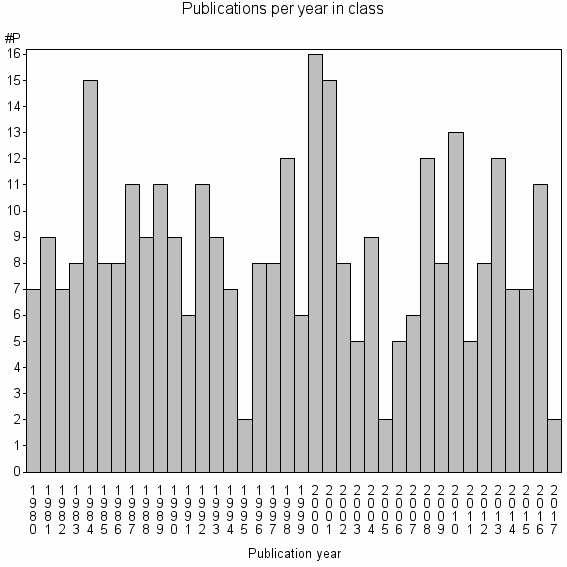
Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
| 24120 | 322 | 35.3 | 66% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 7 | 4 | INFECTIOUS DISEASES//MICROBIOLOGY//VIROLOGY | 1353914 |
| 33 | 3 | JOURNAL OF BACTERIOLOGY//MICROBIOLOGY//MOLECULAR MICROBIOLOGY | 107033 |
| 804 | 2 | JOURNAL OF BACTERIOLOGY//PHOSPHOTRANSFERASE SYSTEM//PERIPLASMIC BINDING PROTEIN | 12049 |
| 24120 | 1 | ARAC PROTEIN//ARAC//ARAC XYLS FAMILY | 322 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | ARAC PROTEIN | authKW | 474149 | 2% | 100% | 5 |
| 2 | ARAC | authKW | 380610 | 5% | 24% | 17 |
| 3 | ARAC XYLS FAMILY | authKW | 379319 | 1% | 100% | 4 |
| 4 | PEPTIDYL ARM | authKW | 213365 | 1% | 75% | 3 |
| 5 | ARM DOMAIN INTERACTIONS | authKW | 189660 | 1% | 100% | 2 |
| 6 | DAHMS PATHWAY | authKW | 189660 | 1% | 100% | 2 |
| 7 | INGN SYST BIOL PROC LISBP UMR792 | address | 189660 | 1% | 100% | 2 |
| 8 | RHABAD | authKW | 189660 | 1% | 100% | 2 |
| 9 | RHAS | authKW | 189660 | 1% | 100% | 2 |
| 10 | ARABINOSE OPERON | authKW | 126438 | 1% | 67% | 2 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Microbiology | 3434 | 43% | 0% | 139 |
| 2 | Biochemistry & Molecular Biology | 955 | 46% | 0% | 148 |
| 3 | Genetics & Heredity | 120 | 9% | 0% | 30 |
| 4 | Biotechnology & Applied Microbiology | 105 | 9% | 0% | 29 |
| 5 | Biophysics | 57 | 6% | 0% | 19 |
| 6 | Biochemical Research Methods | 17 | 3% | 0% | 11 |
| 7 | Biology | 5 | 2% | 0% | 6 |
| 8 | Mathematical & Computational Biology | 1 | 1% | 0% | 2 |
| 9 | Cell Biology | 1 | 3% | 0% | 10 |
| 10 | Integrative & Complementary Medicine | 1 | 0% | 0% | 1 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | INGN SYST BIOL PROC LISBP UMR792 | 189660 | 1% | 100% | 2 |
| 2 | BIOTECHNOLSCI LIFE | 94830 | 0% | 100% | 1 |
| 3 | DUMC DURHAM | 94830 | 0% | 100% | 1 |
| 4 | ESTAC EXPTL ZAIDIN TO 419 | 94830 | 0% | 100% | 1 |
| 5 | ISREC BIOINFORMAT GRP | 94830 | 0% | 100% | 1 |
| 6 | MOLEC CELL BIOL MICROBIOL VIROL | 94830 | 0% | 100% | 1 |
| 7 | PROF ALBAREDA 1 | 94830 | 0% | 100% | 1 |
| 8 | TRINITY MED CLINICAL MICROBIOL | 94830 | 0% | 100% | 1 |
| 9 | BIOCHEM MOL CELLULAR BIOL PLANTS | 81508 | 2% | 12% | 7 |
| 10 | SINTEF MAT CHEM | 48937 | 1% | 13% | 4 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | JOURNAL OF BACTERIOLOGY | 20774 | 26% | 0% | 85 |
| 2 | JOURNAL OF MOLECULAR BIOLOGY | 3644 | 9% | 0% | 30 |
| 3 | ACS SYNTHETIC BIOLOGY | 1526 | 1% | 1% | 3 |
| 4 | GENE | 1234 | 5% | 0% | 16 |
| 5 | PROTEINS-STRUCTURE FUNCTION AND BIOINFORMATICS | 1190 | 3% | 0% | 9 |
| 6 | METABOLIC ENGINEERING | 797 | 1% | 0% | 3 |
| 7 | MOLECULAR MICROBIOLOGY | 784 | 3% | 0% | 10 |
| 8 | MICROBIAL BIOTECHNOLOGY | 638 | 1% | 0% | 2 |
| 9 | NUCLEIC ACIDS RESEARCH | 593 | 5% | 0% | 16 |
| 10 | BIOENGINEERED | 347 | 0% | 0% | 1 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | ARAC PROTEIN | 474149 | 2% | 100% | 5 | Search ARAC+PROTEIN | Search ARAC+PROTEIN |
| 2 | ARAC | 380610 | 5% | 24% | 17 | Search ARAC | Search ARAC |
| 3 | ARAC XYLS FAMILY | 379319 | 1% | 100% | 4 | Search ARAC+XYLS+FAMILY | Search ARAC+XYLS+FAMILY |
| 4 | PEPTIDYL ARM | 213365 | 1% | 75% | 3 | Search PEPTIDYL+ARM | Search PEPTIDYL+ARM |
| 5 | ARM DOMAIN INTERACTIONS | 189660 | 1% | 100% | 2 | Search ARM+DOMAIN+INTERACTIONS | Search ARM+DOMAIN+INTERACTIONS |
| 6 | DAHMS PATHWAY | 189660 | 1% | 100% | 2 | Search DAHMS+PATHWAY | Search DAHMS+PATHWAY |
| 7 | RHABAD | 189660 | 1% | 100% | 2 | Search RHABAD | Search RHABAD |
| 8 | RHAS | 189660 | 1% | 100% | 2 | Search RHAS | Search RHAS |
| 9 | ARABINOSE OPERON | 126438 | 1% | 67% | 2 | Search ARABINOSE+OPERON | Search ARABINOSE+OPERON |
| 10 | ARAC FAMILY | 121921 | 1% | 43% | 3 | Search ARAC+FAMILY | Search ARAC+FAMILY |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | SCHLEIF, R , (2010) ARAC PROTEIN, REGULATION OF THE L-ARABINOSE OPERON IN ESCHERICHIA COLI, AND THE LIGHT SWITCH MECHANISM OF ARAC ACTION.FEMS MICROBIOLOGY REVIEWS. VOL. 34. ISSUE 5. P. 779 -796 | 50 | 71% | 71 |
| 2 | SEEDORFF, J , SCHLEIF, R , (2011) ACTIVE ROLE OF THE INTERDOMAIN LINKER OF ARAC.JOURNAL OF BACTERIOLOGY. VOL. 193. ISSUE 20. P. 5737 -5746 | 30 | 88% | 5 |
| 3 | WICKSTRUM, JR , SKREDENSKE, JM , KOLIN, A , JIN, DJ , FANG, J , EGAN, SM , (2007) TRANSCRIPTION ACTIVATION BY THE DNA-BINDING DOMAIN OF THE ARAC FAMILY PROTEIN RHAS IN THE ABSENCE OF ITS EFFECTOR-BINDING DOMAIN.JOURNAL OF BACTERIOLOGY. VOL. 189. ISSUE 14. P. 4984-4993 | 32 | 74% | 16 |
| 4 | KOLIN, A , BALASUBRAMANIAM, V , SKREDENSKE, JM , WICKSTRUM, JR , EGAN, SM , (2008) DIFFERENCES IN THE MECHANISM OF THE ALLOSTERIC L-RHAMNOSE RESPONSES OF THE ARAC/XYLS FAMILY TRANSCRIPTION ACTIVATORS RHAS AND RHAR.MOLECULAR MICROBIOLOGY. VOL. 68. ISSUE 2. P. 448 -461 | 37 | 63% | 7 |
| 5 | RODGERS, ME , SCHLEIF, R , (2012) HETERODIMERS REVEAL THAT TWO ARABINOSE MOLECULES ARE REQUIRED FOR THE NORMAL ARABINOSE RESPONSE OF ARAC.BIOCHEMISTRY. VOL. 51. ISSUE 41. P. 8085 -8091 | 25 | 78% | 0 |
| 6 | SCHLEIF, R , (2003) ARAC PROTEIN: A LOVE-HATE RELATIONSHIP.BIOESSAYS. VOL. 25. ISSUE 3. P. 274-282 | 25 | 83% | 73 |
| 7 | DIRLA, S , CHIEN, JYH , SCHLEIF, R , (2009) CONSTITUTIVE MUTATIONS IN THE ESCHERICHIA COLI ARAC PROTEIN.JOURNAL OF BACTERIOLOGY. VOL. 191. ISSUE 8. P. 2668-2674 | 21 | 91% | 6 |
| 8 | KAHRAMANOGLOU, C , WEBSTER, CL , EL-ROBH, MS , BELYAEVA, TA , BUSBY, SJW , (2006) MUTATIONAL ANALYSIS OF THE ESCHERICHIA COLI MELR GENE SUGGESTS A TWO-STATE CONCERTED MODEL TO EXPLAIN TRANSCRIPTIONAL ACTIVATION AND REPRESSION IN THE MELIBIOSE OPERON.JOURNAL OF BACTERIOLOGY. VOL. 188. ISSUE 9. P. 3199-3207 | 26 | 79% | 11 |
| 9 | RODGERS, ME , SCHLEIF, R , (2008) DNA TAPE MEASUREMENTS OF ARAC.NUCLEIC ACIDS RESEARCH. VOL. 36. ISSUE 2. P. 404 -410 | 19 | 95% | 1 |
| 10 | DOMINGUEZ-CUEVAS, P , MARIN, P , MARQUES, S , RAMOS, JL , (2008) XV1S-PM PROMOTER INTERACTIONS THROUGH TWO HELIX-TURN-HELIX MOTIFS: IDENTIFYING XY1S RESIDUES IMPORTANT FOR DNA BINDING AND ACTIVATION.JOURNAL OF MOLECULAR BIOLOGY. VOL. 375. ISSUE 1. P. 59-69 | 31 | 55% | 8 |
Classes with closest relation at Level 1 |