Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
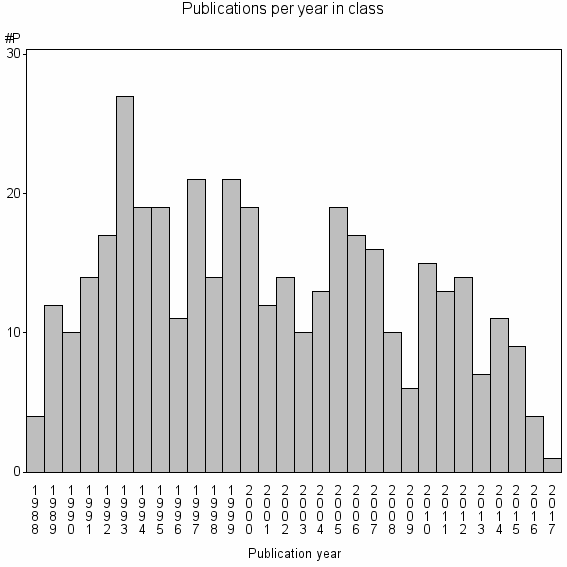
| 21956 | 399 | 19.7 | 80% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 7 | 4 | INFECTIOUS DISEASES//MICROBIOLOGY//VIROLOGY | 1353914 |
| 33 | 3 | JOURNAL OF BACTERIOLOGY//MICROBIOLOGY//MOLECULAR MICROBIOLOGY | 107033 |
| 1609 | 2 | RESTRICTION ENDONUCLEASE//BIOTECHNIQUES//RESTRICTION MODIFICATION SYSTEM | 7100 |
| 21956 | 1 | GENOME WALKING//FALSE POSITIVE EXCLUSION//UNKNOWN FLANKING SEQUENCES | 399 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | GENOME WALKING | authKW | 351826 | 5% | 23% | 20 |
| 2 | FALSE POSITIVE EXCLUSION | authKW | 229587 | 1% | 100% | 3 |
| 3 | UNKNOWN FLANKING SEQUENCES | authKW | 229587 | 1% | 100% | 3 |
| 4 | FLANKING DNA SEQUENCES | authKW | 153058 | 1% | 100% | 2 |
| 5 | FLANKING SEQUENCE AMPLIFICATION | authKW | 153058 | 1% | 100% | 2 |
| 6 | SINGLE PRIMER PCR CORRECTION | authKW | 153058 | 1% | 100% | 2 |
| 7 | UNEVEN PCR | authKW | 153058 | 1% | 100% | 2 |
| 8 | UNIVERSAL FAST WALKING | authKW | 153058 | 1% | 100% | 2 |
| 9 | SINGLE PRIMER PCR | authKW | 122441 | 1% | 40% | 4 |
| 10 | CASSETTE LIGATION MEDIATED PCR | authKW | 102037 | 1% | 67% | 2 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Biochemical Research Methods | 5862 | 40% | 0% | 161 |
| 2 | Biochemistry & Molecular Biology | 2684 | 66% | 0% | 265 |
| 3 | Biotechnology & Applied Microbiology | 607 | 18% | 0% | 70 |
| 4 | Genetics & Heredity | 467 | 15% | 0% | 61 |
| 5 | Chemistry, Analytical | 39 | 6% | 0% | 23 |
| 6 | Plant Sciences | 27 | 5% | 0% | 21 |
| 7 | Microbiology | 24 | 5% | 0% | 19 |
| 8 | Evolutionary Biology | 23 | 2% | 0% | 8 |
| 9 | Cell Biology | 20 | 6% | 0% | 23 |
| 10 | Biophysics | 8 | 3% | 0% | 11 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | BERNHARD NOCHT STRASSE 74 | 76529 | 0% | 100% | 1 |
| 2 | BIOTECHNOL INITIAT FO TRY FO T PROD | 76529 | 0% | 100% | 1 |
| 3 | COMMITTEES EVOLUTIONARY BIOL | 76529 | 0% | 100% | 1 |
| 4 | CYRUS TANG HEMATOL SIP | 76529 | 0% | 100% | 1 |
| 5 | FOODBORNE DIARRHEAL DIS BRANCH MSCO3 | 76529 | 0% | 100% | 1 |
| 6 | FUNCT GENOM TECHNOL TEAMTSURUMI KU | 76529 | 0% | 100% | 1 |
| 7 | INSAUCBUMR 5122 | 76529 | 0% | 100% | 1 |
| 8 | JIANGSU BIO ERSITY BIO OURCES | 76529 | 0% | 100% | 1 |
| 9 | LIFE SCI SUPPORT OFF | 76529 | 0% | 100% | 1 |
| 10 | MACHINE LEARNING DATA MINING BIOINFORMAT | 76529 | 0% | 100% | 1 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | BIOTECHNIQUES | 78959 | 20% | 1% | 79 |
| 2 | METHODS-A COMPANION TO METHODS IN ENZYMOLOGY | 21104 | 1% | 7% | 4 |
| 3 | PCR-METHODS AND APPLICATIONS | 11656 | 1% | 3% | 5 |
| 4 | NUCLEIC ACIDS RESEARCH | 6279 | 14% | 0% | 57 |
| 5 | TRANSGENIC RESEARCH | 3705 | 2% | 1% | 9 |
| 6 | PLANT MOLECULAR BIOLOGY REPORTER | 3528 | 2% | 1% | 8 |
| 7 | MOLECULAR BIOTECHNOLOGY | 2636 | 2% | 0% | 8 |
| 8 | ANALYTICAL BIOCHEMISTRY | 2172 | 6% | 0% | 23 |
| 9 | DNA RESEARCH | 1917 | 1% | 1% | 4 |
| 10 | METHODS IN MOLECULAR AND CELLULAR BIOLOGY | 1232 | 0% | 2% | 1 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | GENOME WALKING | 351826 | 5% | 23% | 20 | Search GENOME+WALKING | Search GENOME+WALKING |
| 2 | FALSE POSITIVE EXCLUSION | 229587 | 1% | 100% | 3 | Search FALSE+POSITIVE+EXCLUSION | Search FALSE+POSITIVE+EXCLUSION |
| 3 | UNKNOWN FLANKING SEQUENCES | 229587 | 1% | 100% | 3 | Search UNKNOWN+FLANKING+SEQUENCES | Search UNKNOWN+FLANKING+SEQUENCES |
| 4 | FLANKING DNA SEQUENCES | 153058 | 1% | 100% | 2 | Search FLANKING+DNA+SEQUENCES | Search FLANKING+DNA+SEQUENCES |
| 5 | FLANKING SEQUENCE AMPLIFICATION | 153058 | 1% | 100% | 2 | Search FLANKING+SEQUENCE+AMPLIFICATION | Search FLANKING+SEQUENCE+AMPLIFICATION |
| 6 | SINGLE PRIMER PCR CORRECTION | 153058 | 1% | 100% | 2 | Search SINGLE+PRIMER+PCR+CORRECTION | Search SINGLE+PRIMER+PCR+CORRECTION |
| 7 | UNEVEN PCR | 153058 | 1% | 100% | 2 | Search UNEVEN+PCR | Search UNEVEN+PCR |
| 8 | UNIVERSAL FAST WALKING | 153058 | 1% | 100% | 2 | Search UNIVERSAL+FAST+WALKING | Search UNIVERSAL+FAST+WALKING |
| 9 | SINGLE PRIMER PCR | 122441 | 1% | 40% | 4 | Search SINGLE+PRIMER+PCR | Search SINGLE+PRIMER+PCR |
| 10 | CASSETTE LIGATION MEDIATED PCR | 102037 | 1% | 67% | 2 | Search CASSETTE+LIGATION+MEDIATED+PCR | Search CASSETTE+LIGATION+MEDIATED+PCR |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | TONOOKA, Y , FUJISHIMA, M , (2009) COMPARISON AND CRITICAL EVALUATION OF PCR-MEDIATED METHODS TO WALK ALONG THE SEQUENCE OF GENOMIC DNA.APPLIED MICROBIOLOGY AND BIOTECHNOLOGY. VOL. 85. ISSUE 1. P. 37 -43 | 28 | 72% | 18 |
| 2 | LI, HX , DING, DQ , CAO, YS , YU, B , GUO, L , LIU, XH , (2015) PARTIALLY OVERLAPPING PRIMER-BASED PCR FOR GENOME WALKING.PLOS ONE. VOL. 10. ISSUE 3. P. - | 16 | 89% | 1 |
| 3 | REDDY, PS , MAHANTY, S , KAUL, T , NAIR, S , SOPORY, SK , REDDY, MK , (2008) A HIGH-THROUGHPUT GENOME-WALKING METHOD AND ITS USE FOR CLONING UNKNOWN FLANKING SEQUENCES.ANALYTICAL BIOCHEMISTRY. VOL. 381. ISSUE 2. P. 248-253 | 19 | 79% | 24 |
| 4 | TRINH, Q , XU, WT , SHI, H , LUO, YB , HUANG, KL , (2012) AN A-T LINKER ADAPTER POLYMERASE CHAIN REACTION METHOD FOR CHROMOSOME WALKING WITHOUT RESTRICTION SITE CLONING BIAS.ANALYTICAL BIOCHEMISTRY. VOL. 425. ISSUE 1. P. 62-67 | 19 | 68% | 3 |
| 5 | YAN, YX , AN, CC , LI, L , GU, JY , TAN, GH , CHEN, ZL , (2003) T-LINKER-SPECIFIC LIGATION PCR (T-LINKER PCR): AN ADVANCED PCR TECHNIQUE FOR CHROMOSOME WALKING OR FOR ISOLATION OF TAGGED DNA ENDS.NUCLEIC ACIDS RESEARCH. VOL. 31. ISSUE 12. P. - | 20 | 77% | 13 |
| 6 | JIANG, Y , PEI, JJ , SONG, X , SHAO, WL , (2007) RESTRICTION SITE-DEPENDENT PCR: AN EFFICIENT TECHNIQUE FOR FAST CLONING OF NEW GENES OF MICROORGANISMS.DNA RESEARCH. VOL. 14. ISSUE 6. P. 283-290 | 18 | 72% | 5 |
| 7 | GADKAR, VJ , FILION, M , (2011) A NOVEL METHOD TO PERFORM GENOMIC WALKS USING A COMBINATION OF SINGLE STRAND DNA CIRCULARIZATION AND ROLLING CIRCLE AMPLIFICATION.JOURNAL OF MICROBIOLOGICAL METHODS. VOL. 87. ISSUE 1. P. 38-43 | 18 | 64% | 1 |
| 8 | XU, P , HU, RY , DING, XY , (2006) OPTIMIZED ADAPTOR POLYMERASE CHAIN REACTION METHOD FOR EFFICIENT GENOMIC WALKING.ACTA BIOCHIMICA ET BIOPHYSICA SINICA. VOL. 38. ISSUE 8. P. 571-576 | 15 | 83% | 3 |
| 9 | LEONI, C , VOLPICELLA, M , DE LEO, F , GALLERANI, R , CECI, LR , (2011) GENOME WALKING IN EUKARYOTES.FEBS JOURNAL. VOL. 278. ISSUE 21. P. 3953 -3977 | 48 | 22% | 16 |
| 10 | BAE, JH , SOHN, JH , (2010) TEMPLATE-BLOCKING PCR: AN ADVANCED PCR TECHNIQUE FOR GENOME WALKING.ANALYTICAL BIOCHEMISTRY. VOL. 398. ISSUE 1. P. 112-116 | 14 | 78% | 10 |
Classes with closest relation at Level 1 |