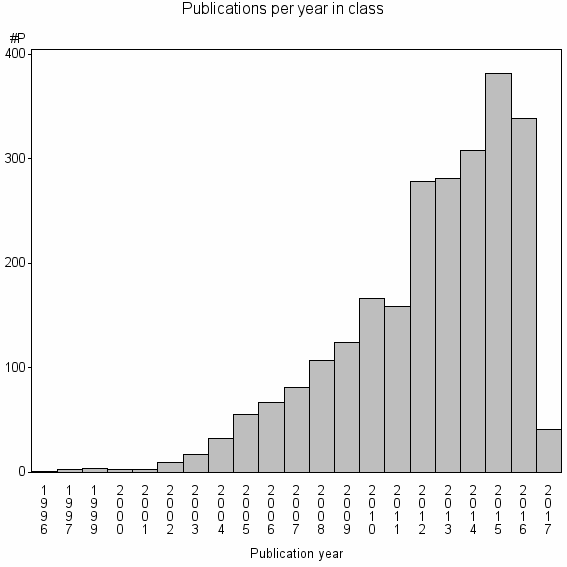
Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
| 1855 | 2456 | 58.1 | 93% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 3 | 4 | PLANT SCIENCES//AGRONOMY//FOOD SCIENCE & TECHNOLOGY | 1882812 |
| 7 | 3 | PLANT SCIENCES//PLANT PHYSIOLOGY//ARABIDOPSIS | 149859 |
| 135 | 2 | PLANT SCIENCES//ARABIDOPSIS//PLANT CELL | 23819 |
| 1855 | 1 | MIR156//MICRORNA//MIRNA | 2456 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | MIR156 | authKW | 425451 | 2% | 93% | 37 |
| 2 | MICRORNA | authKW | 385390 | 26% | 5% | 628 |
| 3 | MIRNA | authKW | 349736 | 15% | 8% | 374 |
| 4 | TASIRNA | authKW | 295326 | 1% | 85% | 28 |
| 5 | DEGRADOME | authKW | 289257 | 2% | 48% | 48 |
| 6 | SMALL RNA | authKW | 248727 | 5% | 17% | 116 |
| 7 | PHASIRNA | authKW | 223761 | 1% | 100% | 18 |
| 8 | MIR172 | authKW | 193399 | 1% | 68% | 23 |
| 9 | MIR396 | authKW | 186467 | 1% | 100% | 15 |
| 10 | TRANS ACTING SIRNA | authKW | 174811 | 1% | 94% | 15 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Plant Sciences | 35436 | 52% | 0% | 1288 |
| 2 | Genetics & Heredity | 5777 | 21% | 0% | 516 |
| 3 | Biochemistry & Molecular Biology | 4328 | 37% | 0% | 901 |
| 4 | Biotechnology & Applied Microbiology | 3030 | 16% | 0% | 392 |
| 5 | Cell Biology | 2725 | 18% | 0% | 454 |
| 6 | Developmental Biology | 480 | 3% | 0% | 78 |
| 7 | Mathematical & Computational Biology | 187 | 2% | 0% | 47 |
| 8 | Biochemical Research Methods | 160 | 4% | 0% | 90 |
| 9 | Horticulture | 100 | 1% | 0% | 32 |
| 10 | Biology | 34 | 2% | 0% | 44 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | INTEGRAT GENOME BIOL | 104458 | 2% | 14% | 60 |
| 2 | PAULOWNIA | 81202 | 1% | 47% | 14 |
| 3 | PLANT BIOL PHD PROGRAM | 76139 | 0% | 88% | 7 |
| 4 | GENE SUPP S | 61193 | 0% | 62% | 8 |
| 5 | DELAWARE BIOTECHNOL | 59235 | 2% | 9% | 51 |
| 6 | CHEMGEN IGERT PROGRAM | 44394 | 0% | 71% | 5 |
| 7 | OASIS ECOL AGR BINTUAN | 37293 | 0% | 100% | 3 |
| 8 | CROP GENOM GENET IMPROVEMENT MOA | 34709 | 0% | 31% | 9 |
| 9 | MED PLANT CULTIVAT | 34527 | 0% | 56% | 5 |
| 10 | IBR BIOL MOL CELULAR ROSARIO | 30589 | 0% | 31% | 8 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | BMC PLANT BIOLOGY | 41236 | 4% | 4% | 88 |
| 2 | FUNCTIONAL & INTEGRATIVE GENOMICS | 22917 | 1% | 6% | 30 |
| 3 | FRONTIERS IN PLANT SCIENCE | 20252 | 4% | 2% | 90 |
| 4 | BMC GENOMICS | 17046 | 5% | 1% | 115 |
| 5 | PLANT CELL | 14604 | 3% | 1% | 84 |
| 6 | PLANT BIOTECHNOLOGY JOURNAL | 11014 | 1% | 3% | 33 |
| 7 | PLANT JOURNAL | 10629 | 3% | 1% | 79 |
| 8 | PLANT MOLECULAR BIOLOGY REPORTER | 6505 | 1% | 2% | 27 |
| 9 | PLANTA | 5006 | 2% | 1% | 60 |
| 10 | JOURNAL OF EXPERIMENTAL BOTANY | 4602 | 3% | 1% | 62 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | MIR156 | 425451 | 2% | 93% | 37 | Search MIR156 | Search MIR156 |
| 2 | MICRORNA | 385390 | 26% | 5% | 628 | Search MICRORNA | Search MICRORNA |
| 3 | MIRNA | 349736 | 15% | 8% | 374 | Search MIRNA | Search MIRNA |
| 4 | TASIRNA | 295326 | 1% | 85% | 28 | Search TASIRNA | Search TASIRNA |
| 5 | DEGRADOME | 289257 | 2% | 48% | 48 | Search DEGRADOME | Search DEGRADOME |
| 6 | SMALL RNA | 248727 | 5% | 17% | 116 | Search SMALL+RNA | Search SMALL+RNA |
| 7 | PHASIRNA | 223761 | 1% | 100% | 18 | Search PHASIRNA | Search PHASIRNA |
| 8 | MIR172 | 193399 | 1% | 68% | 23 | Search MIR172 | Search MIR172 |
| 9 | MIR396 | 186467 | 1% | 100% | 15 | Search MIR396 | Search MIR396 |
| 10 | TRANS ACTING SIRNA | 174811 | 1% | 94% | 15 | Search TRANS+ACTING+SIRNA | Search TRANS+ACTING+SIRNA |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | ZHANG, BH , WANG, QL , (2015) MICRORNA-BASED BIOTECHNOLOGY FOR PLANT IMPROVEMENT.JOURNAL OF CELLULAR PHYSIOLOGY. VOL. 230. ISSUE 1. P. 1 -15 | 163 | 70% | 21 |
| 2 | KHRAIWESH, B , ZHU, JK , ZHU, JH , (2012) ROLE OF MIRNAS AND SIRNAS IN BIOTIC AND ABIOTIC STRESS RESPONSES OF PLANTS.BIOCHIMICA ET BIOPHYSICA ACTA-GENE REGULATORY MECHANISMS. VOL. 1819. ISSUE 2. P. 137 -148 | 102 | 77% | 260 |
| 3 | XIE, M , ZHANG, SX , YU, B , (2015) MICRORNA BIOGENESIS, DEGRADATION AND ACTIVITY IN PLANTS.CELLULAR AND MOLECULAR LIFE SCIENCES. VOL. 72. ISSUE 1. P. 87 -99 | 123 | 78% | 16 |
| 4 | FERDOUS, J , HUSSAIN, SS , SHI, BJ , (2015) ROLE OF MICRORNAS IN PLANT DROUGHT TOLERANCE.PLANT BIOTECHNOLOGY JOURNAL. VOL. 13. ISSUE 3. P. 293 -305 | 130 | 71% | 15 |
| 5 | ELDEM, V , OKAY, S , UNVER, T , (2013) PLANT MICRORNAS: NEW PLAYERS IN FUNCTIONAL GENOMICS.TURKISH JOURNAL OF AGRICULTURE AND FORESTRY. VOL. 37. ISSUE 1. P. 1-21 | 145 | 70% | 26 |
| 6 | TRIPATHI, A , GOSWAMI, K , SANAN-MISHRA, N , (2015) ROLE OF BIOINFORMATICS IN ESTABLISHING MICRORNAS AS MODULATORS OF ABIOTIC STRESS RESPONSES: THE NEW REVOLUTION.FRONTIERS IN PHYSIOLOGY. VOL. 6. ISSUE . P. - | 151 | 66% | 4 |
| 7 | ROGERS, K , CHEN, XM , (2013) BIOGENESIS, TURNOVER, AND MODE OF ACTION OF PLANT MICRORNAS.PLANT CELL. VOL. 25. ISSUE 7. P. 2383 -2399 | 115 | 55% | 168 |
| 8 | VOINNET, O , (2009) ORIGIN, BIOGENESIS, AND ACTIVITY OF PLANT MICRORNAS.CELL. VOL. 136. ISSUE 4. P. 669-687 | 74 | 67% | 885 |
| 9 | LI, C , ZHANG, BH , (2016) MICRORNAS IN CONTROL OF PLANT DEVELOPMENT.JOURNAL OF CELLULAR PHYSIOLOGY. VOL. 231. ISSUE 2. P. 303 -313 | 103 | 53% | 13 |
| 10 | SHRIRAM, V , KUMAR, V , DEVARUMATH, RM , KHARE, TS , WANI, SH , (2016) MICRORNAS AS POTENTIAL TARGETS FOR ABIOTIC STRESS TOLERANCE IN PLANTS.FRONTIERS IN PLANT SCIENCE. VOL. 7. ISSUE . P. - | 118 | 75% | 1 |
Classes with closest relation at Level 1 |