Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
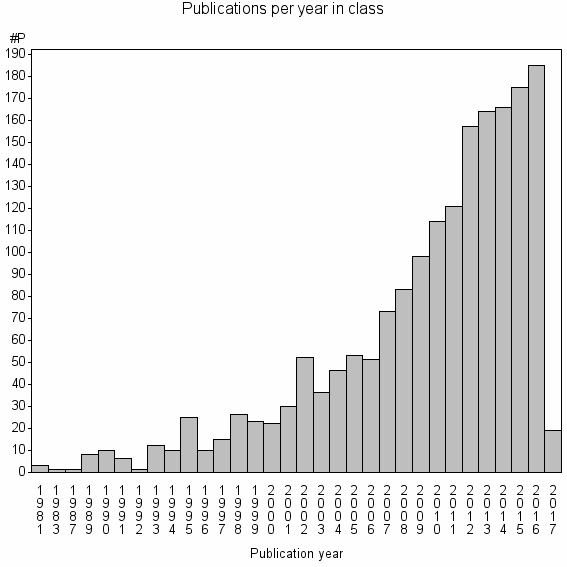
| 4373 | 1796 | 53.5 | 89% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 3 | 4 | PLANT SCIENCES//AGRONOMY//FOOD SCIENCE & TECHNOLOGY | 1882812 |
| 7 | 3 | PLANT SCIENCES//PLANT PHYSIOLOGY//ARABIDOPSIS | 149859 |
| 135 | 2 | PLANT SCIENCES//ARABIDOPSIS//PLANT CELL | 23819 |
| 4373 | 1 | MSAP//RDDM//RNA DIRECTED DNA METHYLATION | 1796 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | MSAP | authKW | 943649 | 4% | 76% | 73 |
| 2 | RDDM | authKW | 418888 | 2% | 79% | 31 |
| 3 | RNA DIRECTED DNA METHYLATION | authKW | 405941 | 2% | 60% | 40 |
| 4 | DNA METHYLATION | authKW | 383791 | 26% | 5% | 467 |
| 5 | GREGOR MENDEL MOL PLANT BIOL | address | 243751 | 2% | 34% | 42 |
| 6 | EPIALLELE | authKW | 214185 | 1% | 60% | 21 |
| 7 | DDM1 | authKW | 207232 | 1% | 76% | 16 |
| 8 | TRANSCRIPTIONAL GENE SILENCING | authKW | 204666 | 2% | 35% | 34 |
| 9 | CYTOSINE METHYLATION | authKW | 179507 | 2% | 27% | 39 |
| 10 | EPIGENETIC VARIATION | authKW | 169981 | 1% | 50% | 20 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Plant Sciences | 18319 | 44% | 0% | 797 |
| 2 | Genetics & Heredity | 11385 | 33% | 0% | 601 |
| 3 | Biochemistry & Molecular Biology | 3757 | 39% | 0% | 709 |
| 4 | Cell Biology | 3440 | 24% | 0% | 423 |
| 5 | Evolutionary Biology | 611 | 4% | 0% | 76 |
| 6 | Horticulture | 394 | 3% | 0% | 48 |
| 7 | Biotechnology & Applied Microbiology | 337 | 7% | 0% | 130 |
| 8 | Developmental Biology | 247 | 3% | 0% | 49 |
| 9 | Agronomy | 111 | 3% | 0% | 49 |
| 10 | Biology | 106 | 3% | 0% | 52 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | GREGOR MENDEL MOL PLANT BIOL | 243751 | 2% | 34% | 42 |
| 2 | SHANGHAI PLANT ST S BIOL | 105351 | 2% | 20% | 31 |
| 3 | INTEGRATED GENET | 79911 | 2% | 17% | 27 |
| 4 | LIFE SCI CORE CURRICULUM | 76498 | 0% | 75% | 6 |
| 5 | MOL EPIGENET MOE | 70161 | 1% | 24% | 17 |
| 6 | MOL CELL DEV BIOL | 67289 | 5% | 5% | 86 |
| 7 | PLANT EPIGENET UNIT | 60712 | 0% | 71% | 5 |
| 8 | DPTO BOS | 43834 | 0% | 37% | 7 |
| 9 | UMR232 | 38249 | 0% | 75% | 3 |
| 10 | PLANT MOL EPIGENET | 38243 | 0% | 38% | 6 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | CURRENT OPINION IN PLANT BIOLOGY | 23202 | 3% | 3% | 49 |
| 2 | PLOS GENETICS | 11483 | 4% | 1% | 64 |
| 3 | PLANT JOURNAL | 7565 | 3% | 1% | 57 |
| 4 | PLANT CELL | 6776 | 3% | 1% | 49 |
| 5 | MOLECULAR PLANT | 6356 | 1% | 2% | 19 |
| 6 | NATURE PLANTS | 6310 | 0% | 5% | 8 |
| 7 | FRONTIERS IN PLANT SCIENCE | 4156 | 2% | 1% | 35 |
| 8 | GENETICS | 4094 | 3% | 0% | 56 |
| 9 | PLANT MOLECULAR BIOLOGY | 3054 | 2% | 1% | 35 |
| 10 | PLANT CELL REPORTS | 2616 | 2% | 1% | 29 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | MSAP | 943649 | 4% | 76% | 73 | Search MSAP | Search MSAP |
| 2 | RDDM | 418888 | 2% | 79% | 31 | Search RDDM | Search RDDM |
| 3 | RNA DIRECTED DNA METHYLATION | 405941 | 2% | 60% | 40 | Search RNA+DIRECTED+DNA+METHYLATION | Search RNA+DIRECTED+DNA+METHYLATION |
| 4 | DNA METHYLATION | 383791 | 26% | 5% | 467 | Search DNA+METHYLATION | Search DNA+METHYLATION |
| 5 | EPIALLELE | 214185 | 1% | 60% | 21 | Search EPIALLELE | Search EPIALLELE |
| 6 | DDM1 | 207232 | 1% | 76% | 16 | Search DDM1 | Search DDM1 |
| 7 | TRANSCRIPTIONAL GENE SILENCING | 204666 | 2% | 35% | 34 | Search TRANSCRIPTIONAL+GENE+SILENCING | Search TRANSCRIPTIONAL+GENE+SILENCING |
| 8 | CYTOSINE METHYLATION | 179507 | 2% | 27% | 39 | Search CYTOSINE+METHYLATION | Search CYTOSINE+METHYLATION |
| 9 | EPIGENETIC VARIATION | 169981 | 1% | 50% | 20 | Search EPIGENETIC+VARIATION | Search EPIGENETIC+VARIATION |
| 10 | METHYLATION SENSITIVE AMPLIFIED POLYMORPHISM MSAP | 154545 | 1% | 91% | 10 | Search METHYLATION+SENSITIVE+AMPLIFIED+POLYMORPHISM+MSAP | Search METHYLATION+SENSITIVE+AMPLIFIED+POLYMORPHISM+MSAP |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | MATZKE, MA , MOSHER, RA , (2014) RNA-DIRECTED DNA METHYLATION: AN EPIGENETIC PATHWAY OF INCREASING COMPLEXITY.NATURE REVIEWS GENETICS. VOL. 15. ISSUE 6. P. 394 -408 | 111 | 74% | 185 |
| 2 | QUADRANA, L , COLOT, V , (2016) PLANT TRANSGENERATIONAL EPIGENETICS.ANNUAL REVIEW OF GENETICS, VOL 50. VOL. 50. ISSUE . P. 467 -491 | 127 | 85% | 0 |
| 3 | KUMAR, S , KUMARI, R , SHARMA, V , SHARMA, V , (2013) ROLES, AND ESTABLISHMENT, MAINTENANCE AND ERASING OF THE EPIGENETIC CYTOSINE METHYLATION MARKS IN PLANTS.JOURNAL OF GENETICS. VOL. 92. ISSUE 3. P. 629 -666 | 178 | 63% | 2 |
| 4 | MATZKE, MA , KANNO, T , MATZKE, AJM , (2015) RNA-DIRECTED DNA METHYLATION: THE EVOLUTION OF A COMPLEX EPIGENETIC PATHWAY IN FLOWERING PLANTS.ANNUAL REVIEW OF PLANT BIOLOGY, VOL 66. VOL. 66. ISSUE . P. 243 -267 | 85 | 62% | 34 |
| 5 | WENDTE, JM , PIKAARD, CS , (2017) THE RNAS OF RNA-DIRECTED DNA METHYLATION.BIOCHIMICA ET BIOPHYSICA ACTA-GENE REGULATORY MECHANISMS. VOL. 1860. ISSUE 1. P. 140 -148 | 87 | 84% | 0 |
| 6 | ZHOU, M , LAW, JA , (2015) RNA POL IV AND V IN GENE SILENCING: REBEL POLYMERASES EVOLVING AWAY FROM POL II'S RULES.CURRENT OPINION IN PLANT BIOLOGY. VOL. 27. ISSUE . P. 154 -164 | 85 | 84% | 2 |
| 7 | EICHTEN, SR , SCHMITZ, RJ , SPRINGER, NM , (2014) EPIGENETICS: BEYOND CHROMATIN MODIFICATIONS AND COMPLEX GENETIC REGULATION.PLANT PHYSIOLOGY. VOL. 165. ISSUE 3. P. 933 -947 | 113 | 58% | 14 |
| 8 | RIGAL, M , BECKER, C , PELISSIER, T , POGORELCNIK, R , DEVOS, J , IKEDA, Y , WEIGEL, D , MATHIEU, O , (2016) EPIGENOME CONFRONTATION TRIGGERS IMMEDIATE REPROGRAMMING OF DNA METHYLATION AND TRANSPOSON SILENCING IN ARABIDOPSIS THALIANA F1 EPIHYBRIDS.PROCEEDINGS OF THE NATIONAL ACADEMY OF SCIENCES OF THE UNITED STATES OF AMERICA. VOL. 113. ISSUE 14. P. E2083 -E2092 | 57 | 89% | 5 |
| 9 | HAUSER, MT , AUFSATZ, W , JONAK, C , LUSCHNIG, C , (2011) TRANSGENERATIONAL EPIGENETIC INHERITANCE IN PLANTS.BIOCHIMICA ET BIOPHYSICA ACTA-GENE REGULATORY MECHANISMS. VOL. 1809. ISSUE 8. P. 459 -468 | 110 | 53% | 86 |
| 10 | VIDALIS, A , ZIVKOVIC, D , WARDENAAR, R , ROQUIS, D , TELLIER, A , JOHANNES, F , (2016) METHYLOME EVOLUTION IN PLANTS.GENOME BIOLOGY. VOL. 17. ISSUE . P. - | 89 | 75% | 0 |
Classes with closest relation at Level 1 |