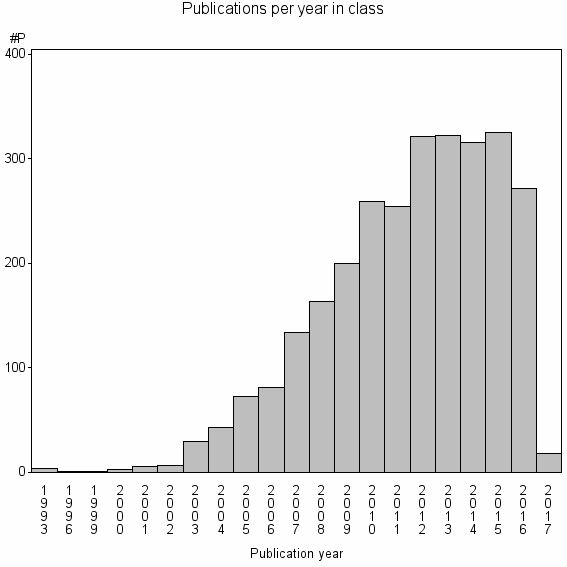
Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
| 1139 | 2827 | 53.0 | 92% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 227 | 3 | MICRORNA//MIRNA//GENE DELIVERY | 49432 |
| 57 | 2 | MICRORNA//MIRNA//LONG NONCODING RNAS | 29594 |
| 1139 | 1 | MICRORNA//MIRNA//DROSHA | 2827 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | MICRORNA | authKW | 358552 | 23% | 5% | 650 |
| 2 | MIRNA | authKW | 205888 | 11% | 6% | 308 |
| 3 | DROSHA | authKW | 180002 | 2% | 34% | 49 |
| 4 | ISOMIR | authKW | 172768 | 1% | 57% | 28 |
| 5 | TARGET PREDICTION | authKW | 114487 | 1% | 31% | 34 |
| 6 | HOST GENE | authKW | 92141 | 1% | 53% | 16 |
| 7 | MICRORNA TARGETS | authKW | 89155 | 1% | 49% | 17 |
| 8 | SILKWORM MULBERRY GENET IMPROVEMENT | address | 79342 | 0% | 57% | 13 |
| 9 | ARM SWITCHING | authKW | 75596 | 0% | 100% | 7 |
| 10 | ARGONAUTE | authKW | 73164 | 2% | 15% | 46 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Mathematical & Computational Biology | 13108 | 13% | 0% | 359 |
| 2 | Genetics & Heredity | 10265 | 26% | 0% | 727 |
| 3 | Biochemistry & Molecular Biology | 10042 | 50% | 0% | 1407 |
| 4 | Biotechnology & Applied Microbiology | 6271 | 21% | 0% | 589 |
| 5 | Cell Biology | 4324 | 21% | 0% | 602 |
| 6 | Biochemical Research Methods | 3075 | 12% | 0% | 332 |
| 7 | Developmental Biology | 1371 | 5% | 0% | 135 |
| 8 | Evolutionary Biology | 251 | 2% | 0% | 67 |
| 9 | Biology | 228 | 3% | 0% | 92 |
| 10 | Biophysics | 159 | 4% | 0% | 109 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | SILKWORM MULBERRY GENET IMPROVEMENT | 79342 | 0% | 57% | 13 |
| 2 | BIOINFORMAT SCI TECHNOL | 42957 | 2% | 9% | 43 |
| 3 | DIANA | 38389 | 0% | 44% | 8 |
| 4 | COMP SCI MATH SCI ENGN | 32398 | 0% | 100% | 3 |
| 5 | SIEE | 32398 | 0% | 100% | 3 |
| 6 | STRATEG TRANSLAT CANC | 28796 | 0% | 67% | 4 |
| 7 | MOE MOL CARDIOL | 24681 | 0% | 57% | 4 |
| 8 | JIANGSU SERICULTURAL BIOL BIOTECHNOL | 24297 | 0% | 75% | 3 |
| 9 | STRATEG CANC PROGRAM | 24297 | 0% | 75% | 3 |
| 10 | GENET CELULAR EMOL | 21599 | 0% | 100% | 2 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | RNA | 35159 | 3% | 4% | 91 |
| 2 | BMC GENOMICS | 22281 | 5% | 1% | 141 |
| 3 | NUCLEIC ACIDS RESEARCH | 14740 | 8% | 1% | 234 |
| 4 | RNA BIOLOGY | 13549 | 1% | 4% | 34 |
| 5 | BIOINFORMATICS | 9996 | 4% | 1% | 100 |
| 6 | BMC BIOINFORMATICS | 7765 | 3% | 1% | 74 |
| 7 | GENOME RESEARCH | 5677 | 1% | 2% | 27 |
| 8 | BMC SYSTEMS BIOLOGY | 5613 | 1% | 2% | 29 |
| 9 | CURRENT BIOINFORMATICS | 5469 | 1% | 3% | 15 |
| 10 | WILEY INTERDISCIPLINARY REVIEWS-RNA | 4214 | 0% | 3% | 12 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | MICRORNA | 358552 | 23% | 5% | 650 | Search MICRORNA | Search MICRORNA |
| 2 | MIRNA | 205888 | 11% | 6% | 308 | Search MIRNA | Search MIRNA |
| 3 | DROSHA | 180002 | 2% | 34% | 49 | Search DROSHA | Search DROSHA |
| 4 | ISOMIR | 172768 | 1% | 57% | 28 | Search ISOMIR | Search ISOMIR |
| 5 | TARGET PREDICTION | 114487 | 1% | 31% | 34 | Search TARGET+PREDICTION | Search TARGET+PREDICTION |
| 6 | HOST GENE | 92141 | 1% | 53% | 16 | Search HOST+GENE | Search HOST+GENE |
| 7 | MICRORNA TARGETS | 89155 | 1% | 49% | 17 | Search MICRORNA+TARGETS | Search MICRORNA+TARGETS |
| 8 | ARM SWITCHING | 75596 | 0% | 100% | 7 | Search ARM+SWITCHING | Search ARM+SWITCHING |
| 9 | ARGONAUTE | 73164 | 2% | 15% | 46 | Search ARGONAUTE | Search ARGONAUTE |
| 10 | MIRTRON | 67489 | 0% | 63% | 10 | Search MIRTRON | Search MIRTRON |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | BARTEL, DP , (2009) MICRORNAS: TARGET RECOGNITION AND REGULATORY FUNCTIONS.CELL. VOL. 136. ISSUE 2. P. 215-233 | 64 | 60% | 7168 |
| 2 | BEREZIKOV, E , (2011) EVOLUTION OF MICRORNA DIVERSITY AND REGULATION IN ANIMALS.NATURE REVIEWS GENETICS. VOL. 12. ISSUE 12. P. 846 -860 | 117 | 73% | 258 |
| 3 | HA, M , KIM, VN , (2014) REGULATION OF MICRORNA BIOGENESIS.NATURE REVIEWS MOLECULAR CELL BIOLOGY. VOL. 15. ISSUE 8. P. 509 -524 | 103 | 38% | 577 |
| 4 | AKHTAR, MM , MICOLUCCI, L , ISLAM, MS , OLIVIERI, F , PROCOPIO, AD , (2016) BIOINFORMATIC TOOLS FOR MICRORNA DISSECTION.NUCLEIC ACIDS RESEARCH. VOL. 44. ISSUE 1. P. 24 -44 | 116 | 67% | 10 |
| 5 | AGARWAL, V , BELL, GW , NAM, JW , BARTEL, DP , (2015) PREDICTING EFFECTIVE MICRORNA TARGET SITES IN MAMMALIAN MRNAS.ELIFE. VOL. 4. ISSUE . P. - | 66 | 58% | 198 |
| 6 | STEINKRAUS, BR , TOEGEL, M , FULGA, TA , (2016) TINY GIANTS OF GENE REGULATION: EXPERIMENTAL STRATEGIES FOR MICRORNA FUNCTIONAL STUDIES.WILEY INTERDISCIPLINARY REVIEWS-DEVELOPMENTAL BIOLOGY. VOL. 5. ISSUE 3. P. 311 -362 | 168 | 53% | 2 |
| 7 | AMERES, SL , ZAMORE, PD , (2013) DIVERSIFYING MICRORNA SEQUENCE AND FUNCTION.NATURE REVIEWS MOLECULAR CELL BIOLOGY. VOL. 14. ISSUE 8. P. 475-488 | 107 | 45% | 312 |
| 8 | HAUSSER, J , ZAVOLAN, M , (2014) IDENTIFICATION AND CONSEQUENCES OF MIRNA-TARGET INTERACTIONS - BEYOND REPRESSION OF GENE EXPRESSION.NATURE REVIEWS GENETICS. VOL. 15. ISSUE 9. P. 599 -612 | 84 | 64% | 105 |
| 9 | CHAWLA, G , SOKOL, NS , (2011) MICRORNAS IN DROSOPHILA DEVELOPMENT.INTERNATIONAL REVIEW OF CELL AND MOLECULAR BIOLOGY, VOL 286. VOL. 286. ISSUE . P. 1 -65 | 155 | 51% | 27 |
| 10 | CHOU, CH , CHANG, NW , SHRESTHA, S , HSU, SD , LIN, YL , LEE, WH , YANG, CD , HONG, HC , WEI, TY , TU, SJ , ET AL (2016) MIRTARBASE 2016: UPDATES TO THE EXPERIMENTALLY VALIDATED MIRNA-TARGET INTERACTIONS DATABASE.NUCLEIC ACIDS RESEARCH. VOL. 44. ISSUE D1. P. D239 -D247 | 37 | 65% | 52 |
Classes with closest relation at Level 1 |