Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
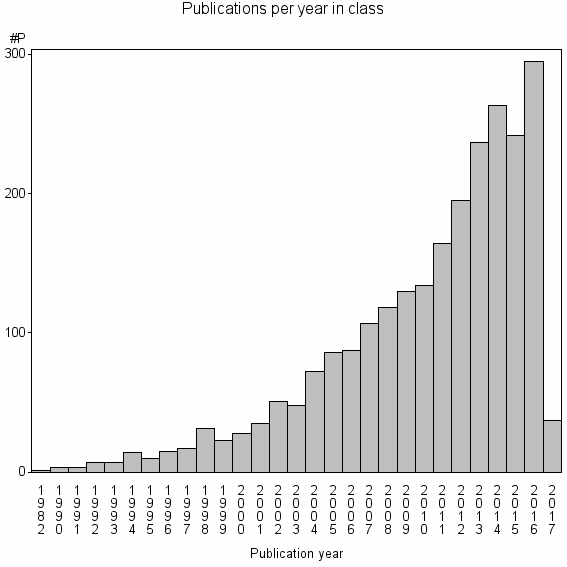
| 1849 | 2460 | 54.8 | 90% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 3 | 4 | PLANT SCIENCES//AGRONOMY//FOOD SCIENCE & TECHNOLOGY | 1882812 |
| 7 | 3 | PLANT SCIENCES//PLANT PHYSIOLOGY//ARABIDOPSIS | 149859 |
| 88 | 2 | PLANT SCIENCES//SALICYLIC ACID//JASMONIC ACID | 26878 |
| 1849 | 1 | NAC TRANSCRIPTION FACTOR//AP2 ERF//ABIOTIC STRESS | 2460 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | NAC TRANSCRIPTION FACTOR | authKW | 568455 | 3% | 68% | 67 |
| 2 | AP2 ERF | authKW | 513755 | 2% | 84% | 49 |
| 3 | ABIOTIC STRESS | authKW | 404312 | 12% | 11% | 296 |
| 4 | DREB | authKW | 395119 | 2% | 86% | 37 |
| 5 | BZIP TRANSCRIPTION FACTOR | authKW | 210798 | 2% | 45% | 38 |
| 6 | ETHYLENE RESPONSE FACTOR | authKW | 204110 | 1% | 66% | 25 |
| 7 | NAC DOMAIN | authKW | 191478 | 1% | 86% | 18 |
| 8 | GCC BOX | authKW | 183855 | 1% | 74% | 20 |
| 9 | DREB TRANSCRIPTION FACTOR | authKW | 137473 | 0% | 92% | 12 |
| 10 | TRANSCRIPTION FACTOR | authKW | 131499 | 14% | 3% | 356 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Plant Sciences | 58075 | 67% | 0% | 1638 |
| 2 | Biochemistry & Molecular Biology | 3033 | 32% | 0% | 777 |
| 3 | Genetics & Heredity | 2294 | 14% | 0% | 340 |
| 4 | Biotechnology & Applied Microbiology | 1438 | 12% | 0% | 283 |
| 5 | Horticulture | 764 | 3% | 0% | 77 |
| 6 | Cell Biology | 360 | 8% | 0% | 202 |
| 7 | Biochemical Research Methods | 207 | 4% | 0% | 99 |
| 8 | Agronomy | 157 | 3% | 0% | 68 |
| 9 | Biophysics | 45 | 3% | 0% | 67 |
| 10 | Agriculture, Multidisciplinary | 23 | 1% | 0% | 26 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | PLANT MOL PHYSIOL | 66726 | 2% | 11% | 49 |
| 2 | SIGNALING PATHWAY UNIT | 63076 | 1% | 27% | 19 |
| 3 | KEY IL CROP GENET OURCES GENET IMPRO | 55280 | 0% | 64% | 7 |
| 4 | DESERT ARID AREAS | 49644 | 0% | 100% | 4 |
| 5 | BIOL OURCES POSTHARVEST | 47397 | 1% | 24% | 16 |
| 6 | PLANT FUNCT GENOM GRP | 39972 | 1% | 17% | 19 |
| 7 | IBADAN STN | 39713 | 0% | 80% | 4 |
| 8 | CROP GERMPLASM OURCES UTILIZATMINIS | 37233 | 0% | 100% | 3 |
| 9 | MOL ANAL GENET IMPROVEMENT | 37233 | 0% | 100% | 3 |
| 10 | STATE AGR BIOL OURCES PROTECT UTIL | 37233 | 0% | 100% | 3 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | PLANT MOLECULAR BIOLOGY REPORTER | 37818 | 3% | 5% | 65 |
| 2 | PLANT MOLECULAR BIOLOGY | 24818 | 5% | 2% | 116 |
| 3 | FUNCTIONAL & INTEGRATIVE GENOMICS | 18527 | 1% | 6% | 27 |
| 4 | FRONTIERS IN PLANT SCIENCE | 14778 | 3% | 2% | 77 |
| 5 | BMC PLANT BIOLOGY | 13785 | 2% | 2% | 51 |
| 6 | PLANT JOURNAL | 13187 | 4% | 1% | 88 |
| 7 | PLANT CELL REPORTS | 10286 | 3% | 1% | 67 |
| 8 | PLANT PHYSIOLOGY AND BIOCHEMISTRY | 7292 | 2% | 1% | 51 |
| 9 | PLANT SCIENCE | 6981 | 3% | 1% | 62 |
| 10 | JOURNAL OF EXPERIMENTAL BOTANY | 6938 | 3% | 1% | 76 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | NAC TRANSCRIPTION FACTOR | 568455 | 3% | 68% | 67 | Search NAC+TRANSCRIPTION+FACTOR | Search NAC+TRANSCRIPTION+FACTOR |
| 2 | AP2 ERF | 513755 | 2% | 84% | 49 | Search AP2+ERF | Search AP2+ERF |
| 3 | ABIOTIC STRESS | 404312 | 12% | 11% | 296 | Search ABIOTIC+STRESS | Search ABIOTIC+STRESS |
| 4 | DREB | 395119 | 2% | 86% | 37 | Search DREB | Search DREB |
| 5 | BZIP TRANSCRIPTION FACTOR | 210798 | 2% | 45% | 38 | Search BZIP+TRANSCRIPTION+FACTOR | Search BZIP+TRANSCRIPTION+FACTOR |
| 6 | ETHYLENE RESPONSE FACTOR | 204110 | 1% | 66% | 25 | Search ETHYLENE+RESPONSE+FACTOR | Search ETHYLENE+RESPONSE+FACTOR |
| 7 | NAC DOMAIN | 191478 | 1% | 86% | 18 | Search NAC+DOMAIN | Search NAC+DOMAIN |
| 8 | GCC BOX | 183855 | 1% | 74% | 20 | Search GCC+BOX | Search GCC+BOX |
| 9 | DREB TRANSCRIPTION FACTOR | 137473 | 0% | 92% | 12 | Search DREB+TRANSCRIPTION+FACTOR | Search DREB+TRANSCRIPTION+FACTOR |
| 10 | TRANSCRIPTION FACTOR | 131499 | 14% | 3% | 356 | Search TRANSCRIPTION+FACTOR | Search TRANSCRIPTION+FACTOR |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | WANG, HY , WANG, HL , SHAO, HB , TANG, XL , (2016) RECENT ADVANCES IN UTILIZING TRANSCRIPTION FACTORS TO IMPROVE PLANT ABIOTIC STRESS TOLERANCE BY TRANSGENIC TECHNOLOGY.FRONTIERS IN PLANT SCIENCE. VOL. 7. ISSUE . P. - | 119 | 69% | 10 |
| 2 | JOSHI, R , WANI, SH , SINGH, B , BOHRA, A , DAR, ZA , LONE, AA , PAREEK, A , SINGLA-PAREEK, SL , (2016) TRANSCRIPTION FACTORS AND PLANTS RESPONSE TO DROUGHT STRESS: CURRENT UNDERSTANDING AND FUTURE DIRECTIONS.FRONTIERS IN PLANT SCIENCE. VOL. 7. ISSUE . P. - | 125 | 71% | 2 |
| 3 | PURANIK, S , SAHU, PP , SRIVASTAVA, PS , PRASAD, M , (2012) NAC PROTEINS: REGULATION AND ROLE IN STRESS TOLERANCE.TRENDS IN PLANT SCIENCE. VOL. 17. ISSUE 6. P. 369 -381 | 73 | 81% | 171 |
| 4 | XU, ZS , CHEN, M , LI, LC , MA, YZ , (2011) FUNCTIONS AND APPLICATION OF THE AP2/ERF TRANSCRIPTION FACTOR FAMILY IN CROP IMPROVEMENT.JOURNAL OF INTEGRATIVE PLANT BIOLOGY. VOL. 53. ISSUE 7. P. 570 -585 | 90 | 66% | 109 |
| 5 | YAMAGUCHI-SHINOZAKI, K , SHINOZAKI, K , (2006) TRANSCRIPTIONAL REGULATORY NETWORKS IN CELLULAR RESPONSES AND TOLERANCE TO DEHYDRATION AND COLD STRESSES.ANNUAL REVIEW OF PLANT BIOLOGY. VOL. 57. ISSUE . P. 781 -803 | 71 | 54% | 1045 |
| 6 | REHMAN, S , MAHMOOD, T , (2015) FUNCTIONAL ROLE OF DREB AND ERF TRANSCRIPTION FACTORS: REGULATING STRESS-RESPONSIVE NETWORK IN PLANTS.ACTA PHYSIOLOGIAE PLANTARUM. VOL. 37. ISSUE 9. P. - | 82 | 78% | 5 |
| 7 | PHUKAN, UJ , JEENA, GS , TRIPATHI, V , SHUKLA, RK , (2017) REGULATION OF APETALA2/ETHYLENE RESPONSE FACTORS IN PLANTS.FRONTIERS IN PLANT SCIENCE. VOL. 8. ISSUE . P. - | 111 | 58% | 0 |
| 8 | LATA, C , PRASAD, M , (2011) ROLE OF DREBS IN REGULATION OF ABIOTIC STRESS RESPONSES IN PLANTS.JOURNAL OF EXPERIMENTAL BOTANY. VOL. 62. ISSUE 14. P. 4731-4748 | 75 | 61% | 159 |
| 9 | AGARWAL, PK , JHA, B , (2010) TRANSCRIPTION FACTORS IN PLANTS AND ABA DEPENDENT AND INDEPENDENT ABIOTIC STRESS SIGNALLING.BIOLOGIA PLANTARUM. VOL. 54. ISSUE 2. P. 201 -212 | 79 | 65% | 104 |
| 10 | LINDEMOSE, S , O'SHEA, C , JENSEN, MK , SKRIVER, K , (2013) STRUCTURE, FUNCTION AND NETWORKS OF TRANSCRIPTION FACTORS INVOLVED IN ABIOTIC STRESS RESPONSES.INTERNATIONAL JOURNAL OF MOLECULAR SCIENCES. VOL. 14. ISSUE 3. P. 5842 -5878 | 94 | 50% | 55 |
Classes with closest relation at Level 1 |