Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
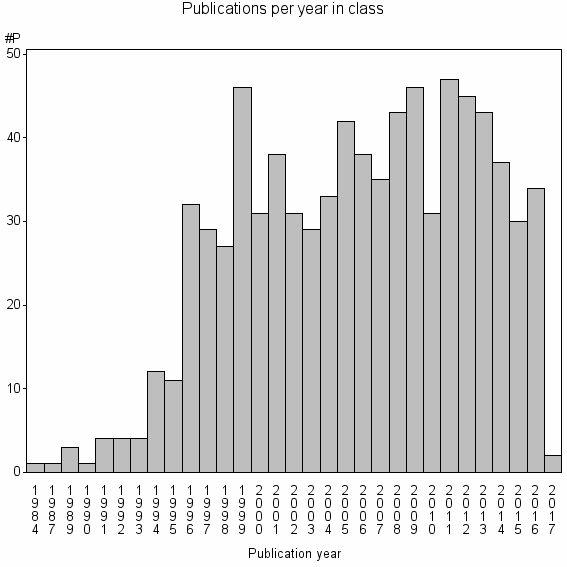
| 13833 | 810 | 49.3 | 93% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 10 | 3 | IMMUNOLOGY//JOURNAL OF IMMUNOLOGY//IMMUNOL | 134299 |
| 741 | 2 | ANTIGEN PRESENTATION//IMMUNOLOGY//ANTIGEN PROCESSING | 12555 |
| 13833 | 1 | PROT CRYSTALLOG UNIT//T CELL RECEPTOR//PEPTIDE MHC | 810 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | PROT CRYSTALLOG UNIT | address | 375383 | 4% | 28% | 35 |
| 2 | T CELL RECEPTOR | authKW | 248104 | 15% | 5% | 124 |
| 3 | PEPTIDE MHC | authKW | 171337 | 1% | 45% | 10 |
| 4 | PEPTIDE MHC COMPLEXES | authKW | 157067 | 1% | 83% | 5 |
| 5 | MOL STAT BIOINFORMAT SECT | address | 150782 | 1% | 67% | 6 |
| 6 | HARPER CANC | address | 119690 | 2% | 19% | 17 |
| 7 | BIOMED COMP SIMULAT BIOINFORMAT | address | 113090 | 0% | 100% | 3 |
| 8 | SECT BIOMED COMP SIMULAT BIOINFORMAT | address | 113090 | 0% | 100% | 3 |
| 9 | T CELL RECEPTOR MIMICS | authKW | 113090 | 0% | 100% | 3 |
| 10 | MOL IMMUNOL SECT | address | 108915 | 2% | 17% | 17 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Immunology | 14976 | 64% | 0% | 521 |
| 2 | Biochemistry & Molecular Biology | 1048 | 32% | 0% | 261 |
| 3 | Biochemical Research Methods | 165 | 6% | 0% | 46 |
| 4 | Medicine, Research & Experimental | 133 | 6% | 0% | 48 |
| 5 | Cell Biology | 102 | 8% | 0% | 63 |
| 6 | Biophysics | 79 | 5% | 0% | 38 |
| 7 | Mathematical & Computational Biology | 26 | 1% | 0% | 11 |
| 8 | Computer Science, Interdisciplinary Applications | 9 | 2% | 0% | 13 |
| 9 | Biotechnology & Applied Microbiology | 5 | 3% | 0% | 22 |
| 10 | Oncology | 0 | 3% | 0% | 22 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | PROT CRYSTALLOG UNIT | 375383 | 4% | 28% | 35 |
| 2 | MOL STAT BIOINFORMAT SECT | 150782 | 1% | 67% | 6 |
| 3 | HARPER CANC | 119690 | 2% | 19% | 17 |
| 4 | BIOMED COMP SIMULAT BIOINFORMAT | 113090 | 0% | 100% | 3 |
| 5 | SECT BIOMED COMP SIMULAT BIOINFORMAT | 113090 | 0% | 100% | 3 |
| 6 | MOL IMMUNOL SECT | 108915 | 2% | 17% | 17 |
| 7 | NEUROIMMUNOL BRANCH | 104131 | 4% | 8% | 33 |
| 8 | PROGRAM BIOMOL STRUCT | 95010 | 1% | 23% | 11 |
| 9 | HUMAN IMMUN | 94236 | 1% | 50% | 5 |
| 10 | SECT BIOSIMULAT BIOINFORMAT | 85669 | 1% | 45% | 5 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | IMMUNITY | 15851 | 5% | 1% | 38 |
| 2 | JOURNAL OF IMMUNOLOGY | 8023 | 13% | 0% | 107 |
| 3 | MOLECULAR IMMUNOLOGY | 7699 | 5% | 1% | 38 |
| 4 | NATURE IMMUNOLOGY | 6142 | 2% | 1% | 19 |
| 5 | EUROPEAN JOURNAL OF IMMUNOLOGY | 5515 | 5% | 0% | 44 |
| 6 | CURRENT OPINION IN IMMUNOLOGY | 3934 | 2% | 1% | 17 |
| 7 | ANNUAL REVIEW OF IMMUNOLOGY | 2783 | 1% | 1% | 8 |
| 8 | JOURNAL OF MOLECULAR BIOLOGY | 2166 | 5% | 0% | 37 |
| 9 | JOURNAL OF EXPERIMENTAL MEDICINE | 1889 | 3% | 0% | 25 |
| 10 | JOURNAL OF IMMUNOLOGICAL METHODS | 1859 | 3% | 0% | 22 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | T CELL RECEPTOR | 248104 | 15% | 5% | 124 | Search T+CELL+RECEPTOR | Search T+CELL+RECEPTOR |
| 2 | PEPTIDE MHC | 171337 | 1% | 45% | 10 | Search PEPTIDE+MHC | Search PEPTIDE+MHC |
| 3 | PEPTIDE MHC COMPLEXES | 157067 | 1% | 83% | 5 | Search PEPTIDE+MHC+COMPLEXES | Search PEPTIDE+MHC+COMPLEXES |
| 4 | T CELL RECEPTOR MIMICS | 113090 | 0% | 100% | 3 | Search T+CELL+RECEPTOR+MIMICS | Search T+CELL+RECEPTOR+MIMICS |
| 5 | V ALPHA DOMAIN | 100521 | 0% | 67% | 4 | Search V+ALPHA+DOMAIN | Search V+ALPHA+DOMAIN |
| 6 | ANTIGEN RECOGNITION | 99919 | 2% | 16% | 17 | Search ANTIGEN+RECOGNITION | Search ANTIGEN+RECOGNITION |
| 7 | PEPTIDE MAJOR HISTOCOMPATIBILITY COMPLEX PMHC | 75393 | 0% | 100% | 2 | Search PEPTIDE+MAJOR+HISTOCOMPATIBILITY+COMPLEX+PMHC | Search PEPTIDE+MAJOR+HISTOCOMPATIBILITY+COMPLEX+PMHC |
| 8 | PEPTIDE MHC CLASS II TCR INTERACTION | 75393 | 0% | 100% | 2 | Search PEPTIDE+MHC+CLASS+II+TCR+INTERACTION | Search PEPTIDE+MHC+CLASS+II+TCR+INTERACTION |
| 9 | TCR MIMICS | 75393 | 0% | 100% | 2 | Search TCR+MIMICS | Search TCR+MIMICS |
| 10 | TCR PMHC | 75393 | 0% | 100% | 2 | Search TCR+PMHC | Search TCR+PMHC |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | ROSSJOHN, J , GRAS, S , MILES, JJ , TURNER, SJ , GODFREY, DI , MCCLUSKEY, J , (2015) T CELL ANTIGEN RECEPTOR RECOGNITION OF ANTIGEN-PRESENTING MOLECULES.ANNUAL REVIEW OF IMMUNOLOGY VOL 33. VOL. 33. ISSUE . P. 169 -200 | 87 | 60% | 43 |
| 2 | ADAMS, JJ , NARAYANAN, S , BIRNBAUM, ME , SIDHU, SS , BLEVINS, SJ , GEE, MH , SIBENER, LV , BAKER, BM , KRANZ, DM , GARCIA, KC , (2016) STRUCTURAL INTERPLAY BETWEEN GERMLINE INTERACTIONS AND ADAPTIVE RECOGNITION DETERMINES THE BANDWIDTH OF TCR-PEPTIDE-MHC CROSS-REACTIVITY.NATURE IMMUNOLOGY. VOL. 17. ISSUE 1. P. 87 -+ | 40 | 78% | 11 |
| 3 | GRAS, S , BURROWS, SR , TURNER, SJ , SEWELL, AK , MCCLUSKEY, J , ROSSJOHN, J , (2012) A STRUCTURAL VOYAGE TOWARD AN UNDERSTANDING OF THE MHC-I-RESTRICTED IMMUNE RESPONSE: LESSONS LEARNED AND MUCH TO BE LEARNED.IMMUNOLOGICAL REVIEWS. VOL. 250. ISSUE . P. 61-81 | 90 | 55% | 25 |
| 4 | BLEVINS, SJ , PIERCE, BG , SINGH, NK , RILEY, TP , WANG, Y , SPEAR, TT , NISHIMURA, MI , WENG, ZP , BAKER, BM , (2016) HOW STRUCTURAL ADAPTABILITY EXISTS ALONGSIDE HLA-A2 BIAS IN THE HUMAN ALPHA BETA TCR REPERTOIRE.PROCEEDINGS OF THE NATIONAL ACADEMY OF SCIENCES OF THE UNITED STATES OF AMERICA. VOL. 113. ISSUE 9. P. E1276 -E1285 | 59 | 74% | 1 |
| 5 | COLE, DK , BULEK, AM , DOLTON, G , SCHAUENBERG, AJ , SZOMOLAY, B , RITTASE, W , TRIMBY, A , JOTHIKUMAR, P , FULLER, A , SKOWERA, A , ET AL (2016) HOTSPOT AUTOIMMUNE T CELL RECEPTOR BINDING UNDERLIES PATHOGEN AND INSULIN PEPTIDE CROSS-REACTIVITY.JOURNAL OF CLINICAL INVESTIGATION. VOL. 126. ISSUE 6. P. 2191 -2204 | 51 | 67% | 5 |
| 6 | BAKER, BM , SCOTT, DR , BLEVINS, SJ , HAWSE, WF , (2012) STRUCTURAL AND DYNAMIC CONTROL OF T-CELL RECEPTOR SPECIFICITY, CROSS-REACTIVITY, AND BINDING MECHANISM.IMMUNOLOGICAL REVIEWS. VOL. 250. ISSUE . P. 10 -31 | 73 | 58% | 21 |
| 7 | BIRNBAUM, ME , MENDOZA, JL , SETHI, DK , DONG, S , GLANVILLE, J , DOBBINS, J , OZKAN, E , DAVIS, MM , WUCHERPFENNIG, KW , GARCIA, KC , (2014) DECONSTRUCTING THE PEPTIDE-MHC SPECIFICITY OF T CELL RECOGNITION.CELL. VOL. 157. ISSUE 5. P. 1073 -1087 | 38 | 58% | 93 |
| 8 | GRAS, S , KJER-NIELSEN, L , CHEN, ZJ , ROSSJOHN, J , MCCLUSKEY, J , (2011) THE STRUCTURAL BASES OF DIRECT T-CELL ALLORECOGNITION: IMPLICATIONS FOR T-CELL-MEDIATED TRANSPLANT REJECTION.IMMUNOLOGY AND CELL BIOLOGY. VOL. 89. ISSUE 3. P. 388-395 | 44 | 85% | 19 |
| 9 | COLE, DK , MILES, KM , MADURA, F , HOLLAND, CJ , SCHAUENBURG, AJA , GODKIN, AJ , BULEK, AM , FULLER, A , AKPOVWA, HJE , PYMM, PG , ET AL (2014) T-CELL RECEPTOR (TCR)-PEPTIDE SPECIFICITY OVERRIDES AFFINITY-ENHANCING TCR-MAJOR HISTOCOMPATIBILITY COMPLEX INTERACTIONS.JOURNAL OF BIOLOGICAL CHEMISTRY. VOL. 289. ISSUE 2. P. 628 -638 | 45 | 75% | 6 |
| 10 | ARMSTRONG, KM , PIEPENBRINK, KH , BAKER, BM , (2008) CONFORMATIONAL CHANGES AND FLEXIBILITY IN T-CELL RECEPTOR RECOGNITION OF PEPTIDE-MHC COMPLEXES.BIOCHEMICAL JOURNAL. VOL. 415. ISSUE . P. 183-196 | 57 | 63% | 44 |
Classes with closest relation at Level 1 |