Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
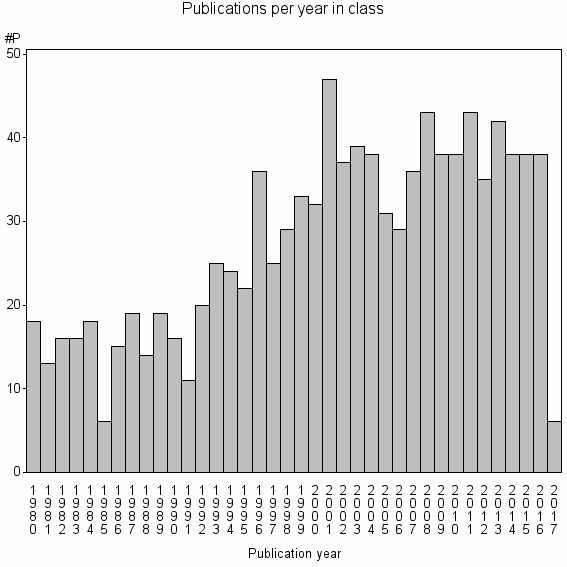
| 10667 | 1043 | 40.7 | 78% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 142 | 3 | DNA REPAIR//INTERNATIONAL JOURNAL OF RADIATION BIOLOGY//RADIATION RESEARCH | 64602 |
| 1228 | 2 | BASE EXCISION REPAIR//DNA GLYCOSYLASE//OXIDATIVE DNA DAMAGE | 9086 |
| 10667 | 1 | URACIL DNA GLYCOSYLASE//DUTPASE//MBD4 | 1043 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | URACIL DNA GLYCOSYLASE | authKW | 1701726 | 9% | 62% | 94 |
| 2 | DUTPASE | authKW | 1521352 | 7% | 73% | 71 |
| 3 | MBD4 | authKW | 296393 | 2% | 56% | 18 |
| 4 | DUTP PYROPHOSPHATASE | authKW | 208175 | 1% | 89% | 8 |
| 5 | DUTP | authKW | 169370 | 1% | 64% | 9 |
| 6 | URACIL DNA GLYCOSYLASE INHIBITOR | authKW | 117100 | 0% | 100% | 4 |
| 7 | UNG | authKW | 112584 | 1% | 38% | 10 |
| 8 | UDG | authKW | 110203 | 1% | 47% | 8 |
| 9 | GENOME METAB REPAIR | address | 93678 | 0% | 80% | 4 |
| 10 | DEOXYURIDINE TRIPHOSPHATE NUCLEOTIDOHYDROLASE | authKW | 87825 | 0% | 100% | 3 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Biochemistry & Molecular Biology | 4894 | 56% | 0% | 588 |
| 2 | Virology | 1046 | 8% | 0% | 84 |
| 3 | Biophysics | 831 | 11% | 0% | 116 |
| 4 | Toxicology | 567 | 8% | 0% | 80 |
| 5 | Genetics & Heredity | 488 | 10% | 0% | 107 |
| 6 | Cell Biology | 292 | 10% | 0% | 109 |
| 7 | Oncology | 172 | 9% | 0% | 91 |
| 8 | Biochemical Research Methods | 161 | 5% | 0% | 53 |
| 9 | Crystallography | 97 | 3% | 0% | 34 |
| 10 | Biotechnology & Applied Microbiology | 91 | 5% | 0% | 57 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | GENOME METAB REPAIR | 93678 | 0% | 80% | 4 |
| 2 | ENZYMOL | 68417 | 5% | 4% | 57 |
| 3 | SOUTH CAROLINA EXPT STN | 65867 | 0% | 75% | 3 |
| 4 | GENOME METAB | 58550 | 0% | 100% | 2 |
| 5 | DOCTORAL MULTIDISCIPLINARY MED SCI | 52692 | 0% | 60% | 3 |
| 6 | PL BIOTECHNOL FOOD SCI | 45518 | 2% | 8% | 19 |
| 7 | RADIOL CHEM BIOMED ENGN | 39032 | 0% | 67% | 2 |
| 8 | BIOCHEM 576 MED SCI BLDG | 29275 | 0% | 100% | 1 |
| 9 | BIOCHEM BIOPHYS MICROBIOL | 29275 | 0% | 100% | 1 |
| 10 | BIOCHEM MOLEC BIOL STRUCT BIOCHEM SECT | 29275 | 0% | 100% | 1 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | DNA REPAIR | 19660 | 4% | 2% | 37 |
| 2 | NUCLEIC ACIDS RESEARCH | 6052 | 9% | 0% | 91 |
| 3 | CURRENT PROTEIN & PEPTIDE SCIENCE | 3255 | 1% | 1% | 10 |
| 4 | MUTATION RESEARCH-DNA REPAIR | 2932 | 1% | 1% | 7 |
| 5 | JOURNAL OF BIOLOGICAL CHEMISTRY | 2092 | 10% | 0% | 107 |
| 6 | ACTA CRYSTALLOGRAPHICA SECTION D-BIOLOGICAL CRYSTALLOGRAPHY | 1472 | 1% | 0% | 15 |
| 7 | PROGRESS IN NUCLEIC ACID RESEARCH AND MOLECULAR BIOLOGY | 1366 | 0% | 1% | 5 |
| 8 | BIOCHEMISTRY | 1046 | 4% | 0% | 45 |
| 9 | MUTATION RESEARCH | 1014 | 2% | 0% | 17 |
| 10 | ACTA CRYSTALLOGRAPHICA SECTION D-STRUCTURAL BIOLOGY | 831 | 1% | 0% | 6 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | URACIL DNA GLYCOSYLASE | 1701726 | 9% | 62% | 94 | Search URACIL+DNA+GLYCOSYLASE | Search URACIL+DNA+GLYCOSYLASE |
| 2 | DUTPASE | 1521352 | 7% | 73% | 71 | Search DUTPASE | Search DUTPASE |
| 3 | MBD4 | 296393 | 2% | 56% | 18 | Search MBD4 | Search MBD4 |
| 4 | DUTP PYROPHOSPHATASE | 208175 | 1% | 89% | 8 | Search DUTP+PYROPHOSPHATASE | Search DUTP+PYROPHOSPHATASE |
| 5 | DUTP | 169370 | 1% | 64% | 9 | Search DUTP | Search DUTP |
| 6 | URACIL DNA GLYCOSYLASE INHIBITOR | 117100 | 0% | 100% | 4 | Search URACIL+DNA+GLYCOSYLASE+INHIBITOR | Search URACIL+DNA+GLYCOSYLASE+INHIBITOR |
| 7 | UNG | 112584 | 1% | 38% | 10 | Search UNG | Search UNG |
| 8 | UDG | 110203 | 1% | 47% | 8 | Search UDG | Search UDG |
| 9 | DEOXYURIDINE TRIPHOSPHATE NUCLEOTIDOHYDROLASE | 87825 | 0% | 100% | 3 | Search DEOXYURIDINE+TRIPHOSPHATE+NUCLEOTIDOHYDROLASE | Search DEOXYURIDINE+TRIPHOSPHATE+NUCLEOTIDOHYDROLASE |
| 10 | THYMIDYLATE DEPRIVATION | 87825 | 0% | 100% | 3 | Search THYMIDYLATE+DEPRIVATION | Search THYMIDYLATE+DEPRIVATION |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | VERTESSY, BG , TOTH, J , (2009) KEEPING URACIL OUT OF DNA: PHYSIOLOGICAL ROLE, STRUCTURE AND CATALYTIC MECHANISM OF DUTPASES.ACCOUNTS OF CHEMICAL RESEARCH. VOL. 42. ISSUE 1. P. 97 -106 | 49 | 88% | 63 |
| 2 | SCHORMANN, N , RICCIARDI, R , CHATTOPADHYAY, D , (2014) URACIL-DNA GLYCOSYLASES-STRUCTURAL AND FUNCTIONAL PERSPECTIVES ON AN ESSENTIAL FAMILY OF DNA REPAIR ENZYMES.PROTEIN SCIENCE. VOL. 23. ISSUE 12. P. 1667 -1685 | 52 | 70% | 15 |
| 3 | NYIRI, K , PAPP-KADAR, V , SZABO, JE , NEMETH, V , VERTESSY, BG , (2015) EXPLORING THE ROLE OF THE PHAGE-SPECIFIC INSERT OF BACTERIOPHAGE I BROKEN VERTICAL BAR 11 DUTPASE.STRUCTURAL CHEMISTRY. VOL. 26. ISSUE 5-6. P. 1425 -1432 | 40 | 93% | 1 |
| 4 | VASILENKO, NL , NEVINSKY, GA , (2003) PATHWAYS OF ACCUMULATION AND REPAIR OF DEOXYURIDINE RESIDUES IN DNA OF HIGHER AND LOWER ORGANISMS.BIOCHEMISTRY-MOSCOW. VOL. 68. ISSUE 2. P. 135-151 | 61 | 76% | 3 |
| 5 | MOSBAUGH, DW , BENNETT, SE , (1994) URACIL-EXCISION DNA-REPAIR.PROGRESS IN NUCLEIC ACID RESEARCH AND MOLECULAR BIOLOGY, VOL 48. VOL. 48. ISSUE . P. 315 -370 | 106 | 53% | 72 |
| 6 | HIZI, A , HERZIG, E , (2015) DUTPASE: THE FREQUENTLY OVERLOOKED ENZYME ENCODED BY MANY RETROVIRUSES.RETROVIROLOGY. VOL. 12. ISSUE . P. - | 54 | 58% | 3 |
| 7 | ZHARKOV, DO , MECHETIN, GV , NEVINSKY, GA , (2010) URACIL-DNA GLYCOSYLASE: STRUCTURAL, THERMODYNAMIC AND KINETIC ASPECTS OF LESION SEARCH AND RECOGNITION.MUTATION RESEARCH-FUNDAMENTAL AND MOLECULAR MECHANISMS OF MUTAGENESIS. VOL. 685. ISSUE 1-2. P. 11 -20 | 53 | 58% | 37 |
| 8 | KROKAN, HE , DRABLOS, F , SLUPPHAUG, G , (2002) URACIL IN DNA - OCCURRENCE, CONSEQUENCES AND REPAIR.ONCOGENE. VOL. 21. ISSUE 58. P. 8935-8948 | 56 | 56% | 211 |
| 9 | HIRMONDO, R , SZABO, JE , NYIRI, K , TARJANYI, S , DOBROTKA, P , TOTH, J , VERTESSY, BG , (2015) CROSS-SPECIES INHIBITION OF DUTPASE VIA THE STAPHYLOCOCCAL STL PROTEIN PERTURBS DNTP POOL AND COLONY FORMATION IN MYCOBACTERIUM.DNA REPAIR. VOL. 30. ISSUE . P. 21 -27 | 38 | 72% | 1 |
| 10 | SZABO, JE , NEMETH, V , PAPP-KADAR, V , NYIRI, K , LEVELES, I , BENDES, AA , ZAGYVA, I , RONA, G , PALINKAS, HL , BESZTERCEI, B , ET AL (2014) HIGHLY POTENT DUTPASE INHIBITION BY A BACTERIAL REPRESSOR PROTEIN REVEALS A NOVEL MECHANISM FOR GENE EXPRESSION CONTROL.NUCLEIC ACIDS RESEARCH. VOL. 42. ISSUE 19. P. 11912 -11920 | 33 | 79% | 4 |
Classes with closest relation at Level 1 |