Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
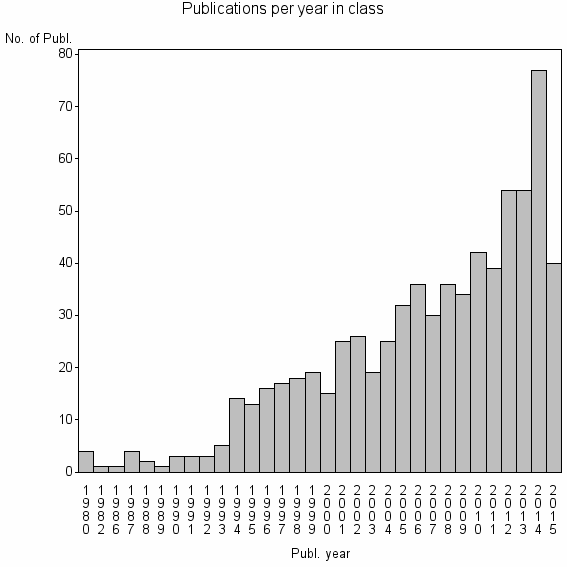
| 14369 | 708 | 44.3 | 89% |
Classes in level above (level 2) |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | TRIGGER FACTOR | Author keyword | 16 | 35% | 5% | 37 |
| 2 | NASCENT CHAIN | Author keyword | 10 | 63% | 1% | 10 |
| 3 | COTRANSLATIONAL FOLDING | Author keyword | 7 | 43% | 2% | 13 |
| 4 | CO TRANSLATIONAL PROTEIN FOLDING | Author keyword | 6 | 80% | 1% | 4 |
| 5 | UPSTREAM OPEN READING FRAME | Author keyword | 6 | 35% | 2% | 15 |
| 6 | NASCENT PEPTIDE | Author keyword | 6 | 44% | 2% | 11 |
| 7 | UORF | Author keyword | 5 | 27% | 2% | 17 |
| 8 | AUG CONTEXT | Author keyword | 5 | 63% | 1% | 5 |
| 9 | EXIT TUNNEL | Author keyword | 5 | 50% | 1% | 7 |
| 10 | NASCENT CHAINS | Author keyword | 4 | 75% | 0% | 3 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | TRIGGER FACTOR | 16 | 35% | 5% | 37 | Search TRIGGER+FACTOR | Search TRIGGER+FACTOR |
| 2 | NASCENT CHAIN | 10 | 63% | 1% | 10 | Search NASCENT+CHAIN | Search NASCENT+CHAIN |
| 3 | COTRANSLATIONAL FOLDING | 7 | 43% | 2% | 13 | Search COTRANSLATIONAL+FOLDING | Search COTRANSLATIONAL+FOLDING |
| 4 | CO TRANSLATIONAL PROTEIN FOLDING | 6 | 80% | 1% | 4 | Search CO+TRANSLATIONAL+PROTEIN+FOLDING | Search CO+TRANSLATIONAL+PROTEIN+FOLDING |
| 5 | UPSTREAM OPEN READING FRAME | 6 | 35% | 2% | 15 | Search UPSTREAM+OPEN+READING+FRAME | Search UPSTREAM+OPEN+READING+FRAME |
| 6 | NASCENT PEPTIDE | 6 | 44% | 2% | 11 | Search NASCENT+PEPTIDE | Search NASCENT+PEPTIDE |
| 7 | UORF | 5 | 27% | 2% | 17 | Search UORF | Search UORF |
| 8 | AUG CONTEXT | 5 | 63% | 1% | 5 | Search AUG+CONTEXT | Search AUG+CONTEXT |
| 9 | EXIT TUNNEL | 5 | 50% | 1% | 7 | Search EXIT+TUNNEL | Search EXIT+TUNNEL |
| 10 | NASCENT CHAINS | 4 | 75% | 0% | 3 | Search NASCENT+CHAINS | Search NASCENT+CHAINS |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | EXIT TUNNEL | 40 | 49% | 8% | 59 |
| 2 | TRANSLATING RIBOSOMES | 35 | 74% | 4% | 26 |
| 3 | TRYPTOPHAN INDUCTION | 18 | 65% | 2% | 17 |
| 4 | ARGININE ATTENUATOR PEPTIDE | 15 | 77% | 1% | 10 |
| 5 | NASCENT POLYPEPTIDE | 9 | 39% | 3% | 19 |
| 6 | NASCENT PEPTIDE | 9 | 24% | 5% | 34 |
| 7 | RIBOSOMAL EXIT TUNNEL | 8 | 52% | 2% | 11 |
| 8 | TNA OPERON EXPRESSION | 8 | 56% | 1% | 10 |
| 9 | PROLYL TRANSFER RNA | 7 | 57% | 1% | 8 |
| 10 | 5 TRANSCRIPT LEADER | 6 | 71% | 1% | 5 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Emerging evidence for functional peptides encoded by short open reading frames | 2014 | 30 | 114 | 45% |
| Identification and characterization of sORF-encoded polypeptides | 2015 | 1 | 58 | 43% |
| Upstream open reading frames as regulators of mRNA translation | 2000 | 398 | 65 | 37% |
| The ribosome as a platform for co-translational processing, folding and targeting of newly synthesized proteins | 2009 | 167 | 118 | 31% |
| Structure and function of the molecular chaperone Trigger Factor | 2010 | 52 | 139 | 48% |
| Divergent stalling sequences sense and control cellular physiology | 2010 | 47 | 55 | 47% |
| Folding at the birth of the nascent chain: coordinating translation with co-translational folding | 2011 | 33 | 49 | 59% |
| Arrest Peptides: Cis-Acting Modulators of Translation | 2013 | 27 | 172 | 41% |
| Ribosome-associated chaperones as key players in proteostasis | 2012 | 39 | 67 | 36% |
| Nascent polypeptide sequences that influence ribosome function | 2011 | 16 | 49 | 71% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | BIOPHYS MOL BIOL GENET | 2 | 16% | 2.0% | 14 |
| 2 | IBGC UMR CNRS 5095 | 2 | 67% | 0.3% | 2 |
| 3 | CELL BIOL INTERNAL MED | 1 | 50% | 0.1% | 1 |
| 4 | CHINA UK | 1 | 50% | 0.1% | 1 |
| 5 | CNRS UPS2682 | 1 | 50% | 0.1% | 1 |
| 6 | FONDAZ CARISBO | 1 | 50% | 0.1% | 1 |
| 7 | JEAN PIERRE BOURGINSACLAY PLANT SCI | 1 | 50% | 0.1% | 1 |
| 8 | MSPRL | 1 | 50% | 0.1% | 1 |
| 9 | RNA SYST BIOL CELLULAR MOL PHARMACOL | 1 | 50% | 0.1% | 1 |
| 10 | WFS | 1 | 50% | 0.1% | 1 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000142365 | TRANS TRANSLATION//TMRNA//SMPB |
| 2 | 0.0000120449 | ERM METHYLTRANSFERASE//CMLA//MLS ANTIBIOTIC RESISTANCE |
| 3 | 0.0000118168 | IRES//INTERNAL RIBOSOME ENTRY SITE IRES//INTERNAL RIBOSOME ENTRY SITE |
| 4 | 0.0000117187 | ALPHA NAC//NASCENT POLYPEPTIDE ASSOCIATED COMPLEX//FIAT |
| 5 | 0.0000106332 | TRANSLATIONAL SELECTION//CODON USAGE//SYNONYMOUS CODON USAGE |
| 6 | 0.0000099027 | SIGNAL RECOGNITION PARTICLE//SRP RECEPTOR//FTSY |
| 7 | 0.0000097774 | 2A PEPTIDE//FMDV 2A//2A |
| 8 | 0.0000092213 | EIF3//GENE REGULAT DEV//EIF1 |
| 9 | 0.0000090193 | ELONGATION FACTOR TU//EF G//ELONGATION FACTOR G |
| 10 | 0.0000086346 | DNAJ//GRPE//DNAK |