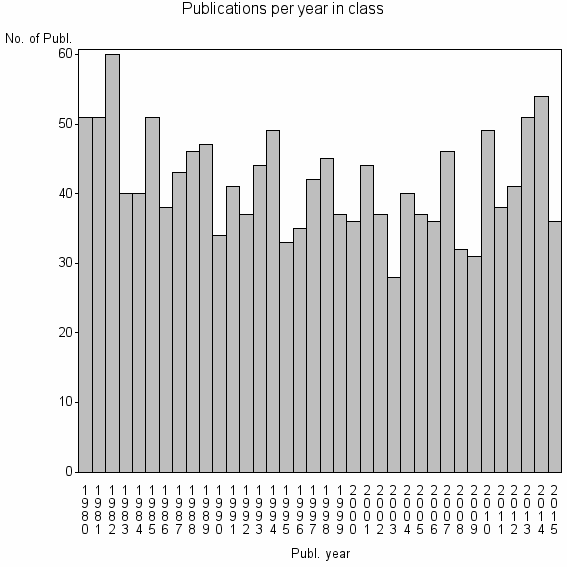
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 5827 | 1500 | 42.6 | 69% |
Classes in level above (level 2) |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | EIF3 | Author keyword | 34 | 59% | 3% | 38 |
| 2 | GENE REGULAT DEV | Address | 26 | 27% | 6% | 83 |
| 3 | EIF1 | Author keyword | 21 | 85% | 1% | 11 |
| 4 | EIF5 | Author keyword | 20 | 100% | 1% | 9 |
| 5 | HEME REGULATED INHIBITOR | Author keyword | 15 | 88% | 0% | 7 |
| 6 | EIF2 | Author keyword | 13 | 36% | 2% | 30 |
| 7 | INT6 | Author keyword | 13 | 80% | 1% | 8 |
| 8 | EIF1A | Author keyword | 10 | 63% | 1% | 10 |
| 9 | INT 6 | Author keyword | 9 | 83% | 0% | 5 |
| 10 | TRANSLATION INITIATION | Author keyword | 9 | 11% | 5% | 77 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | EIF3 | 34 | 59% | 3% | 38 | Search EIF3 | Search EIF3 |
| 2 | EIF1 | 21 | 85% | 1% | 11 | Search EIF1 | Search EIF1 |
| 3 | EIF5 | 20 | 100% | 1% | 9 | Search EIF5 | Search EIF5 |
| 4 | HEME REGULATED INHIBITOR | 15 | 88% | 0% | 7 | Search HEME+REGULATED+INHIBITOR | Search HEME+REGULATED+INHIBITOR |
| 5 | EIF2 | 13 | 36% | 2% | 30 | Search EIF2 | Search EIF2 |
| 6 | INT6 | 13 | 80% | 1% | 8 | Search INT6 | Search INT6 |
| 7 | EIF1A | 10 | 63% | 1% | 10 | Search EIF1A | Search EIF1A |
| 8 | INT 6 | 9 | 83% | 0% | 5 | Search INT+6 | Search INT+6 |
| 9 | TRANSLATION INITIATION | 9 | 11% | 5% | 77 | Search TRANSLATION+INITIATION | Search TRANSLATION+INITIATION |
| 10 | EIF3F | 8 | 75% | 0% | 6 | Search EIF3F | Search EIF3F |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | MULTIFACTOR COMPLEX | 206 | 98% | 3% | 52 |
| 2 | POLYPEPTIDE CHAIN INITIATION | 82 | 65% | 5% | 79 |
| 3 | 40S RIBOSOMAL SUBUNIT | 68 | 79% | 3% | 44 |
| 4 | REVERSING FACTOR | 56 | 89% | 2% | 25 |
| 5 | START CODON SELECTION | 52 | 77% | 2% | 36 |
| 6 | PRT1 PROTEIN | 45 | 90% | 1% | 19 |
| 7 | FACTOR EIF 2 | 37 | 73% | 2% | 29 |
| 8 | EIF 2 ALPHA KINASE | 33 | 51% | 3% | 47 |
| 9 | FACTOR EIF3 | 27 | 78% | 1% | 18 |
| 10 | AUG SELECTION | 26 | 87% | 1% | 13 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| The Scanning Mechanism of Eukaryotic Translation Initiation | 2014 | 32 | 228 | 61% |
| The mechanism of eukaryotic translation initiation and principles of its regulation | 2010 | 570 | 113 | 27% |
| A mechanistic overview of translation initiation in eukaryotes | 2012 | 71 | 106 | 70% |
| Translational regulation of yeast GCN4 - A window on factors that control initiator-tRNA binding to the ribosome | 1997 | 351 | 52 | 92% |
| Translational regulation of GCN4 and the general amino acid control of yeast | 2005 | 481 | 182 | 64% |
| eIF3: a versatile scaffold for translation initiation complexes | 2006 | 184 | 79 | 81% |
| Molecular Mechanism of Scanning and Start Codon Selection in Eukaryotes | 2011 | 79 | 213 | 50% |
| Molecular View of 43 S Complex Formation and Start Site Selection in Eukaryotic Translation Initiation | 2010 | 37 | 49 | 92% |
| TRANSLATIONAL CONTROL OF GCN4 - AN IN-VIVO BAROMETER OF INITIATION-FACTOR ACTIVITY | 1994 | 145 | 32 | 78% |
| 'Ribozoomin' - Translation Initiation from the Perspective of the Ribosome-bound Eukaryotic Initiation Factors (eIFs) | 2012 | 26 | 210 | 54% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | GENE REGULAT DEV | 26 | 27% | 5.5% | 83 |
| 2 | REGULAT GENE EXP S | 7 | 36% | 1.1% | 16 |
| 3 | EUKARYOT GENE REGULAT | 5 | 18% | 1.6% | 24 |
| 4 | MECH REGULAT PROT SYNTH | 3 | 100% | 0.2% | 3 |
| 5 | UNITE MIXTE RECH 7654 | 2 | 33% | 0.3% | 5 |
| 6 | DIBIFIM | 1 | 50% | 0.1% | 2 |
| 7 | DPT BIOTECNOL CELLULARI EMATOL | 1 | 100% | 0.1% | 2 |
| 8 | BIOCHIM CNRS UMR7654 | 1 | 50% | 0.1% | 1 |
| 9 | CANC PHARMACOL TOXICOL | 1 | 50% | 0.1% | 1 |
| 10 | HYDROBIOLSTATE F HWATER ECOL BI | 1 | 50% | 0.1% | 1 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000180810 | EIF4E//EIF 4E//CAPPING EFFICIENCY |
| 2 | 0.0000165934 | PKR//VA RNA//NF90 |
| 3 | 0.0000131787 | IRES//INTERNAL RIBOSOME ENTRY SITE IRES//INTERNAL RIBOSOME ENTRY SITE |
| 4 | 0.0000106857 | ELONGATION FACTOR 2 KINASE//EUKARYOTIC ELONGATION FACTOR 2 EEF2//ELONGATION FACTOR 2 |
| 5 | 0.0000102557 | POLYA BINDING PROTEIN//POLY A BINDING PROTEIN//PABP |
| 6 | 0.0000101292 | EEF1A2//ELONGATION FACTOR 1 GAMMA//ELONGATION FACTOR 1 |
| 7 | 0.0000092213 | TRIGGER FACTOR//NASCENT CHAIN//COTRANSLATIONAL FOLDING |
| 8 | 0.0000066287 | ELONGATION FACTOR TU//EF G//ELONGATION FACTOR G |
| 9 | 0.0000065854 | RNA HELICASE//DEAD BOX//DEAD BOX PROTEIN |
| 10 | 0.0000063774 | S6K2//S6 KINASE//RIBOSOMAL S6 KINASE 2 S6K2 |