The Hober Lab
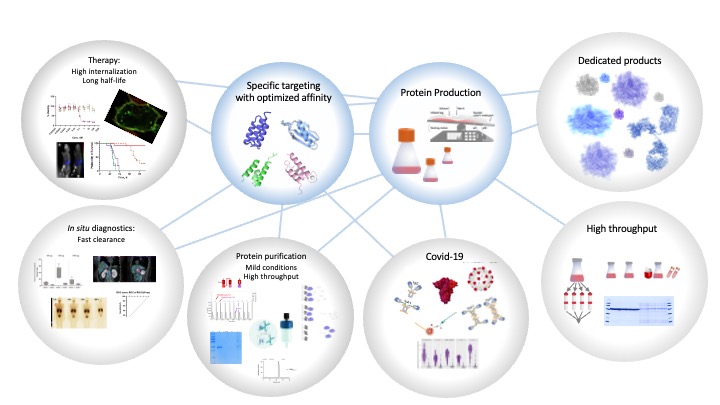
The focus of the research group is to develop predictable and robust systems for protein production, purification and detection. Well-characterized, small and folded domains descending from the bacterial receptors staphylococcal protein A (SPA) or streptococcal protein G (SPG) are used as scaffolds.
Their known three-dimensional structures, a capability of independent folding and the absence of disulfide bridges make these domains ideal as frameworks for further protein engineering. Protein-engineering strategies are used in order to design proteins for protein purification processes.
The goal with the mutagenesis could be either to stabilize or destabilize ligands for different purposes. The designed purification tags and antibodies produced within the HPR program are combined to develop strategies for capture of proteins and protein complexes in an efficient and selective manner. Furthermore, high throughput methods for production and purification of proteins are developed.
Depending on protein and aim we are using either mammalian cells or bacteria for the protein production. The high throughput purification systems are based on different affinity chromatography methods.
During 2020 Hober lab has been very active in the development of new methods for serological analyses of immunological response to SARS-CoV-2. This has been made possible in collaboration with Nilsson, Tegel end Hedhammar. Luminex-based methods for antibody detection and pseudoneutralization has been developed and used to analyze more than 200 000 samples . The methods have been used to follow the Health Care workers at Danderyds hospital, The COMMUNITY and also in several other scientific collaborations.
In the menu to the left, some of our projects are described in more detail.
Dissertations from the group:
- Torbjörn Gräslund (2001) “Protein engineering by directed evolution and rational design” (shared with Prof. Nygren)
- Susanne Gülich (2002) “Engineering of proteinaceous ligands for improved performance in affinity chromatography applications”
- Martin Linhult(2003) “Protein engineering to explore and improve affinity ligands”
- Maria Ehn (2003) “Protein based approaches for further development of the pyrosequencing technology platform”
- My Hedhammar (2005) “Strategies for facilitated protein recovery after recombinant production in Escherishia coli”
- Ronny Falk (2006) “Systems enabling antibody-mediated proteomics research”
- Karin Larsson (2009) “Generation and characterization of antibodies for proteomics research” (shared with Dr. Wernéus)
- Cecilia Eriksson (2009) “Affinity based proteomic research tools”
- Tove Alm (2010) “Interaction-engineered three-helix bundle domains for protein recovery and detection”
- Johanna Steen (2010) “Characterization of antigenic properties and high throughput protein purification” (shared with Doc. Ottosson)
- Anna Konrad (2012 (Lic.)) “Development of a covalent site-specific antibody labeling strategy by the use of photoactivable Z domains”
- Johan Nilvebrant (2012) “An albumin-binding domain as a scaffold for bispecific affinity proteins”
- Hanna Tegel (2013) “Proteom wide protein production”
- Tove Boström (2014) “High-throughput protein analysis using mass spectrometry-based methods”
- Mikael Åstrand (2016) “Engineering strategies for ABD-derived affinity proteins for therapeutic and diagnostic applications”
- Sara Kanje (2016) "Engineering of small IgG binding domains for antibody labelling and purification"
- Sarah Lindbo (2018) "Generation and engineering of ABD-derived proteins for clinical applications"
- Emma von Witting (2020) "The ADAPT scaffold as a tool for diagnostic imaging and targeted therapy"
- Julia Scheffel (2022) "Calcium-dependent affinity domains for the purification of antibodies and antibody fragments"
- Andreas Wisniewski (2023) "Bifunctional ADAPTs: Opportunity for serological Half-life extension and Targeted therapy"
- Malin Jönsson (2023) "Engineering alternative scaffold proteins for conditional targeting"
- Marit Möller (2024) "Engineering conditional binding for enhanced protein therapeutics"
Principal Investigator:
