Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
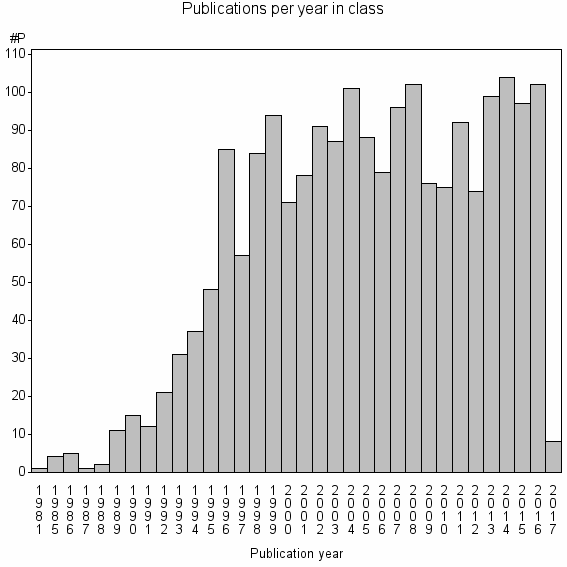
| 3254 | 2028 | 48.8 | 92% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 142 | 3 | DNA REPAIR//INTERNATIONAL JOURNAL OF RADIATION BIOLOGY//RADIATION RESEARCH | 64602 |
| 271 | 2 | INTERNATIONAL JOURNAL OF RADIATION BIOLOGY//RADIATION RESEARCH//ATAXIA TELANGIECTASIA | 19288 |
| 3254 | 1 | DNA PK//NON HOMOLOGOUS END JOINING//DNA DEPENDENT PROTEIN KINASE | 2028 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | DNA PK | authKW | 1218317 | 7% | 55% | 147 |
| 2 | NON HOMOLOGOUS END JOINING | authKW | 869696 | 9% | 33% | 175 |
| 3 | DNA DEPENDENT PROTEIN KINASE | authKW | 836354 | 4% | 67% | 83 |
| 4 | KU | authKW | 683482 | 4% | 58% | 78 |
| 5 | KU70 | authKW | 617265 | 4% | 49% | 83 |
| 6 | DNA PKCS | authKW | 577556 | 4% | 47% | 81 |
| 7 | XRCC4 | authKW | 524545 | 3% | 62% | 56 |
| 8 | KU80 | authKW | 474687 | 3% | 52% | 61 |
| 9 | NHEJ | authKW | 429926 | 4% | 37% | 78 |
| 10 | KU PROTEIN | authKW | 368829 | 2% | 70% | 35 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Biochemistry & Molecular Biology | 5236 | 43% | 0% | 877 |
| 2 | Cell Biology | 4688 | 26% | 0% | 520 |
| 3 | Genetics & Heredity | 3740 | 19% | 0% | 381 |
| 4 | Oncology | 1935 | 18% | 0% | 357 |
| 5 | Toxicology | 1781 | 10% | 0% | 193 |
| 6 | Biology | 903 | 7% | 0% | 134 |
| 7 | Biophysics | 765 | 8% | 0% | 163 |
| 8 | Radiology, Nuclear Medicine & Medical Imaging | 563 | 8% | 0% | 155 |
| 9 | Immunology | 117 | 6% | 0% | 113 |
| 10 | Biotechnology & Applied Microbiology | 91 | 4% | 0% | 89 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | MED RADIAT BIOL | 167611 | 2% | 32% | 35 |
| 2 | UNITE DEV NORMAL PATHOL SYST IMMUNITAIRE | 130114 | 1% | 79% | 11 |
| 3 | DNA REPAIR GENE | 101618 | 0% | 75% | 9 |
| 4 | PROGRAM GENE REGULAT | 87098 | 0% | 64% | 9 |
| 5 | MOL RADIAT BIOL | 79148 | 2% | 16% | 33 |
| 6 | UNIT MED RADIAT BIOL | 60216 | 0% | 67% | 6 |
| 7 | GENOME DAMAGE STABIL | 53030 | 2% | 8% | 45 |
| 8 | HRICHTUNG BIOPHYS | 51901 | 0% | 34% | 10 |
| 9 | GENOME DAMAGE STABIL UNIT | 47043 | 0% | 63% | 5 |
| 10 | PHARM PL SCI | 43968 | 1% | 19% | 15 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | DNA REPAIR | 91122 | 5% | 5% | 111 |
| 2 | MUTATION RESEARCH-DNA REPAIR | 9981 | 1% | 4% | 18 |
| 3 | RADIATION RESEARCH | 7629 | 3% | 1% | 58 |
| 4 | NUCLEIC ACIDS RESEARCH | 4979 | 6% | 0% | 116 |
| 5 | MUTATION RESEARCH-FUNDAMENTAL AND MOLECULAR MECHANISMS OF MUTAGENESIS | 3201 | 1% | 1% | 29 |
| 6 | INTERNATIONAL JOURNAL OF RADIATION BIOLOGY | 2813 | 2% | 1% | 31 |
| 7 | JOURNAL OF RADIATION RESEARCH | 2603 | 1% | 1% | 19 |
| 8 | MOLECULAR AND CELLULAR BIOLOGY | 2265 | 3% | 0% | 59 |
| 9 | JOURNAL OF BIOLOGICAL CHEMISTRY | 1547 | 7% | 0% | 132 |
| 10 | MOLECULAR CANCER RESEARCH | 1339 | 1% | 1% | 13 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | DNA PK | 1218317 | 7% | 55% | 147 | Search DNA+PK | Search DNA+PK |
| 2 | NON HOMOLOGOUS END JOINING | 869696 | 9% | 33% | 175 | Search NON+HOMOLOGOUS+END+JOINING | Search NON+HOMOLOGOUS+END+JOINING |
| 3 | DNA DEPENDENT PROTEIN KINASE | 836354 | 4% | 67% | 83 | Search DNA+DEPENDENT+PROTEIN+KINASE | Search DNA+DEPENDENT+PROTEIN+KINASE |
| 4 | KU | 683482 | 4% | 58% | 78 | Search KU | Search KU |
| 5 | KU70 | 617265 | 4% | 49% | 83 | Search KU70 | Search KU70 |
| 6 | DNA PKCS | 577556 | 4% | 47% | 81 | Search DNA+PKCS | Search DNA+PKCS |
| 7 | XRCC4 | 524545 | 3% | 62% | 56 | Search XRCC4 | Search XRCC4 |
| 8 | KU80 | 474687 | 3% | 52% | 61 | Search KU80 | Search KU80 |
| 9 | NHEJ | 429926 | 4% | 37% | 78 | Search NHEJ | Search NHEJ |
| 10 | KU PROTEIN | 368829 | 2% | 70% | 35 | Search KU+PROTEIN | Search KU+PROTEIN |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | MAHANEY, BL , MEEK, K , LEES-MILLER, SP , (2009) REPAIR OF IONIZING RADIATION-INDUCED DNA DOUBLE-STRAND BREAKS BY NON-HOMOLOGOUS END-JOINING.BIOCHEMICAL JOURNAL. VOL. 417. ISSUE . P. 639 -650 | 114 | 67% | 311 |
| 2 | DAVIS, AJ , CHEN, DJ , (2013) DNA DOUBLE STRAND BREAK REPAIR VIA NON-HOMOLOGOUS END-JOINING.TRANSLATIONAL CANCER RESEARCH. VOL. 2. ISSUE 3. P. 130 -143 | 99 | 80% | 59 |
| 3 | SMITH, GCM , JACKSON, SP , (1999) THE DNA-DEPENDENT PROTEIN KINASE.GENES & DEVELOPMENT. VOL. 13. ISSUE 8. P. 916 -934 | 139 | 62% | 634 |
| 4 | MEEK, K , DANG, V , LEES-MILLER, SP , (2008) DNA-PK: THE MEANS TO JUSTIFY THE ENDS?.ADVANCES IN IMMUNOLOGY, VOL 99. VOL. 99. ISSUE . P. 33-58 | 102 | 88% | 92 |
| 5 | DAVIS, AJ , CHEN, BPC , CHEN, DJ , (2014) DNA-PK: A DYNAMIC ENZYME IN A VERSATILE DSB REPAIR PATHWAY.DNA REPAIR. VOL. 17. ISSUE . P. 21 -29 | 97 | 67% | 44 |
| 6 | TUTEJA, R , TUTEJA, N , (2000) KU AUTOANTIGEN: A MULTIFUNCTIONAL DNA-BINDING PROTEIN.CRITICAL REVIEWS IN BIOCHEMISTRY AND MOLECULAR BIOLOGY. VOL. 35. ISSUE 1. P. 1 -33 | 150 | 71% | 137 |
| 7 | DYNAN, WS , YOO, S , (1998) INTERACTION OF KU PROTEIN AND DNA-DEPENDENT PROTEIN KINASE CATALYTIC SUBUNIT WITH NUCLEIC ACIDS.NUCLEIC ACIDS RESEARCH. VOL. 26. ISSUE 7. P. 1551-1559 | 120 | 83% | 230 |
| 8 | LIEBER, MR , (2010) THE MECHANISM OF DOUBLE-STRAND DNA BREAK REPAIR BY THE NONHOMOLOGOUS DNA END-JOINING PATHWAY.ANNUAL REVIEW OF BIOCHEMISTRY, VOL 79. VOL. 79. ISSUE . P. 181-211 | 77 | 50% | 726 |
| 9 | CHIRUVELLA, KK , LIANG, ZB , WILSON, TE , (2013) REPAIR OF DOUBLE-STRAND BREAKS BY END JOINING.COLD SPRING HARBOR PERSPECTIVES IN BIOLOGY. VOL. 5. ISSUE 5. P. - | 110 | 55% | 67 |
| 10 | JETTE, N , LEES-MILLER, SP , (2015) THE DNA-DEPENDENT PROTEIN KINASE: A MULTIFUNCTIONAL PROTEIN KINASE WITH ROLES IN DNA DOUBLE STRAND BREAK REPAIR AND MITOSIS.PROGRESS IN BIOPHYSICS & MOLECULAR BIOLOGY. VOL. 117. ISSUE 2-3. P. 194 -205 | 91 | 65% | 18 |
Classes with closest relation at Level 1 |