Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
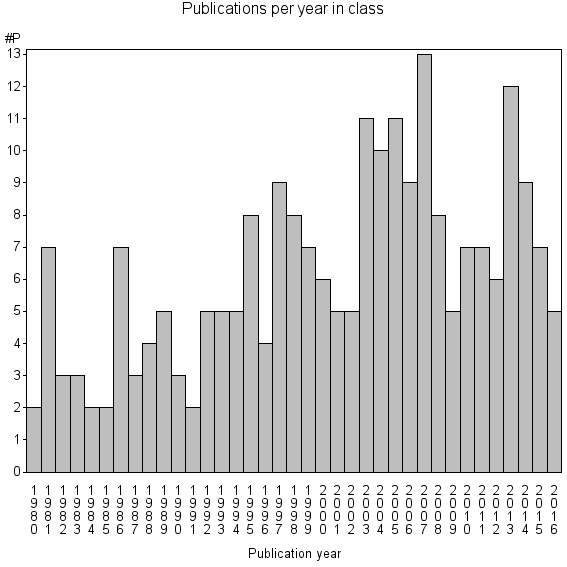
| 27408 | 230 | 34.3 | 66% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 278 | 3 | CORYNEBACTERIUM GLUTAMICUM//ACTA CRYSTALLOGRAPHICA SECTION D-BIOLOGICAL CRYSTALLOGRAPHY//BIOCHEMISTRY & MOLECULAR BIOLOGY | 43163 |
| 1967 | 2 | GLUTAMATE DEHYDROGENASE//3 ISOPROPYLMALATE DEHYDROGENASE//ISOCITRATE DEHYDROGENASE | 5642 |
| 27408 | 1 | HALOPHILIC//HALOPHILIC ADAPTATION//PL MOL MICROBIOL | 230 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | HALOPHILIC | authKW | 630761 | 13% | 16% | 29 |
| 2 | HALOPHILIC ADAPTATION | authKW | 414879 | 2% | 63% | 5 |
| 3 | PL MOL MICROBIOL | address | 321268 | 5% | 22% | 11 |
| 4 | NUCLEOSIDE DIPHOSPHATE KINASE | authKW | 272173 | 10% | 9% | 23 |
| 5 | POLYEXTREME | authKW | 265525 | 1% | 100% | 2 |
| 6 | HALOPHILIC ENZYME | authKW | 250196 | 3% | 27% | 7 |
| 7 | HALOPHILIC PROTEINS | authKW | 199141 | 1% | 50% | 3 |
| 8 | HALOPHILE | authKW | 167731 | 10% | 6% | 22 |
| 9 | HALOFERAX VOLCANII | authKW | 167317 | 5% | 11% | 11 |
| 10 | HALOPHILIC ENZYMES | authKW | 151723 | 2% | 29% | 4 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Biochemistry & Molecular Biology | 1575 | 67% | 0% | 154 |
| 2 | Biophysics | 516 | 18% | 0% | 41 |
| 3 | Microbiology | 280 | 16% | 0% | 36 |
| 4 | Biochemical Research Methods | 159 | 10% | 0% | 22 |
| 5 | Biotechnology & Applied Microbiology | 87 | 10% | 0% | 22 |
| 6 | Crystallography | 80 | 6% | 0% | 13 |
| 7 | Chemistry, Applied | 19 | 3% | 0% | 8 |
| 8 | Cell Biology | 11 | 6% | 0% | 13 |
| 9 | Spectroscopy | 3 | 2% | 0% | 4 |
| 10 | Biology | 3 | 2% | 0% | 4 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | PL MOL MICROBIOL | 321268 | 5% | 22% | 11 |
| 2 | ABT MEMEBRANBIOCHEM | 132762 | 0% | 100% | 1 |
| 3 | BIOL ORGANOMETALL CATALYSIS | 132762 | 0% | 100% | 1 |
| 4 | BIOL STRUCTLBM | 132762 | 0% | 100% | 1 |
| 5 | BIOPHYS MOL BIOL STRUCT J P EBEL | 132762 | 0% | 100% | 1 |
| 6 | BIOPHYS MOL SEIN | 132762 | 0% | 100% | 1 |
| 7 | BIOTEHCNOL CHEM ENGN | 132762 | 0% | 100% | 1 |
| 8 | BMCC INSERM U309 DBMC | 132762 | 0% | 100% | 1 |
| 9 | BURDWAN RAJ | 132762 | 0% | 100% | 1 |
| 10 | CONTRIBUT STRUCT BIOL UNIT | 132762 | 0% | 100% | 1 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | EXTREMOPHILES | 11355 | 5% | 1% | 11 |
| 2 | ANNUAL REVIEW OF BIOPHYSICS AND BIOENGINEERING | 5528 | 1% | 2% | 2 |
| 3 | PROTEIN JOURNAL | 2634 | 2% | 0% | 4 |
| 4 | PROTEIN AND PEPTIDE LETTERS | 2556 | 3% | 0% | 7 |
| 5 | BIOPHYSICAL CHEMISTRY | 1120 | 3% | 0% | 6 |
| 6 | ANNUAL REVIEW OF BIOPHYSICS AND BIOPHYSICAL CHEMISTRY | 974 | 0% | 1% | 1 |
| 7 | ARCHAEA-AN INTERNATIONAL MICROBIOLOGICAL JOURNAL | 895 | 0% | 1% | 1 |
| 8 | FOLDING & DESIGN | 748 | 0% | 1% | 1 |
| 9 | BIOCHEMICAL SOCIETY SYMPOSIA | 716 | 0% | 1% | 1 |
| 10 | JOURNAL OF MOLECULAR BIOLOGY | 675 | 5% | 0% | 11 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | HALOPHILIC | 630761 | 13% | 16% | 29 | Search HALOPHILIC | Search HALOPHILIC |
| 2 | HALOPHILIC ADAPTATION | 414879 | 2% | 63% | 5 | Search HALOPHILIC+ADAPTATION | Search HALOPHILIC+ADAPTATION |
| 3 | NUCLEOSIDE DIPHOSPHATE KINASE | 272173 | 10% | 9% | 23 | Search NUCLEOSIDE+DIPHOSPHATE+KINASE | Search NUCLEOSIDE+DIPHOSPHATE+KINASE |
| 4 | POLYEXTREME | 265525 | 1% | 100% | 2 | Search POLYEXTREME | Search POLYEXTREME |
| 5 | HALOPHILIC ENZYME | 250196 | 3% | 27% | 7 | Search HALOPHILIC+ENZYME | Search HALOPHILIC+ENZYME |
| 6 | HALOPHILIC PROTEINS | 199141 | 1% | 50% | 3 | Search HALOPHILIC+PROTEINS | Search HALOPHILIC+PROTEINS |
| 7 | HALOPHILE | 167731 | 10% | 6% | 22 | Search HALOPHILE | Search HALOPHILE |
| 8 | HALOFERAX VOLCANII | 167317 | 5% | 11% | 11 | Search HALOFERAX+VOLCANII | Search HALOFERAX+VOLCANII |
| 9 | HALOPHILIC ENZYMES | 151723 | 2% | 29% | 4 | Search HALOPHILIC+ENZYMES | Search HALOPHILIC+ENZYMES |
| 10 | ASWAN HIGH DAM LAKE | 132762 | 0% | 100% | 1 | Search ASWAN+HIGH+DAM+LAKE | Search ASWAN+HIGH+DAM+LAKE |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | SINHA, R , KHARE, SK , (2014) PROTECTIVE ROLE OF SALT IN CATALYSIS AND MAINTAINING STRUCTURE OF HALOPHILIC PROTEINS AGAINST DENATURATION.FRONTIERS IN MICROBIOLOGY. VOL. 5. ISSUE . P. - | 30 | 60% | 4 |
| 2 | MADERN, D , EBEL, C , ZACCAI, G , (2000) HALOPHILIC ADAPTATION OF ENZYMES.EXTREMOPHILES. VOL. 4. ISSUE 2. P. 91 -98 | 27 | 51% | 249 |
| 3 | MADERN, D , EBEL, C , (2007) INFLUENCE OF AN ANION-BINDING SITE IN THE STABILIZATION OF HALOPHILIC MALATE DEHYDROGENASE FROM HALOARCULA MARISMORTUI.BIOCHIMIE. VOL. 89. ISSUE 8. P. 981 -987 | 22 | 67% | 8 |
| 4 | GRAZIANO, G , MERLINO, A , (2014) MOLECULAR BASES OF PROTEIN HALOTOLERANCE.BIOCHIMICA ET BIOPHYSICA ACTA-PROTEINS AND PROTEOMICS. VOL. 1844. ISSUE 4. P. 850 -858 | 29 | 38% | 14 |
| 5 | ESCLAPEZ, J , PIRE, C , BAUTISTA, V , MARTINEZ-ESPINOSA, RM , FERRER, J , BONETE, MJ , (2007) ANALYSIS OF ACIDIC SURFACE OF HALOFERAX MEDITERRANEI GLUCOSE DEHYDROGENASE BY SITE-DIRECTED MUTAGENESIS.FEBS LETTERS. VOL. 581. ISSUE 5. P. 837-842 | 17 | 77% | 8 |
| 6 | TOKUNAGA, H , ARAKAWA, T , TOKUNAGA, M , (2008) ENGINEERING OF HALOPHILIC ENZYMES: TWO ACIDIC AMINO ACID RESIDUES AT THE CARBOXY-TERMINAL REGION CONFER HALOPHILIC CHARACTERISTICS TO HALOMONAS AND PSEUDOMONAS NUCLEOSIDE DIPHOSPHATE KINASES.PROTEIN SCIENCE. VOL. 17. ISSUE 9. P. 1603 -1610 | 15 | 75% | 21 |
| 7 | BANDYOPADHYAY, AK , KRISHNAMOORTHY, G , PADHY, LC , SONAWAT, HM , (2007) KINETICS OF SALT-DEPENDENT UNFOLDING OF [2FE-2S] FERREDOXIN OF HALOBACTERIUM SALINARUM.EXTREMOPHILES. VOL. 11. ISSUE 4. P. 615-625 | 19 | 54% | 10 |
| 8 | ARAKAWA, T , TOKUNAGA, M , (2004) ELECTROSTATIC AND HYDROPHOBIC INTERACTIONS PLAY A MAJOR ROLE IN THE STABILITY AND REFOLDING OF HALOPHILIC PROTEINS.PROTEIN AND PEPTIDE LETTERS. VOL. 11. ISSUE 2. P. 125-132 | 18 | 62% | 14 |
| 9 | TOKUNAGA, M , ARAKAWA, T , ISHIBASHI, M , TOKUNAGA, H , (2005) FAMILIAR EXTREMOPHILES, HALOPHILES, AND HALOPHILIC ENZYMES.SEIKAGAKU. VOL. 77. ISSUE 4. P. 320-331 | 22 | 46% | 0 |
| 10 | EBEL, C , COSTENARO, L , PASCU, M , FAOU, P , KERNEL, B , PROUST-DE MARTIN, F , ZACCAI, G , (2002) SOLVENT INTERACTIONS OF HALOPHILIC MALATE DEHYDROGENASE.BIOCHEMISTRY. VOL. 41. ISSUE 44. P. 13234-13244 | 21 | 50% | 31 |
Classes with closest relation at Level 1 |