Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
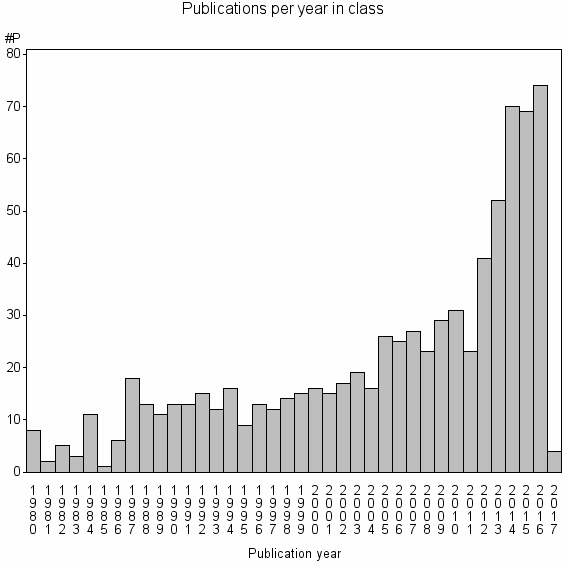
| 14173 | 787 | 46.8 | 85% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 153 | 3 | BIOCHEMISTRY & MOLECULAR BIOLOGY//RIBOSOME//RNA | 62593 |
| 1632 | 2 | EIF4E//TRANSLATION INITIATION//IRES | 6984 |
| 14173 | 1 | RIBOSOME PROFILING//UORF//UPSTREAM OPEN READING FRAME | 787 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | RIBOSOME PROFILING | authKW | 496181 | 3% | 47% | 27 |
| 2 | UORF | authKW | 405536 | 4% | 37% | 28 |
| 3 | UPSTREAM OPEN READING FRAME | authKW | 225715 | 2% | 36% | 16 |
| 4 | SCANNING MODEL | authKW | 190107 | 1% | 70% | 7 |
| 5 | AUG CONTEXT | authKW | 174589 | 1% | 75% | 6 |
| 6 | ARABIDOPSIS THALIANA T87 | authKW | 116395 | 0% | 100% | 3 |
| 7 | KOZAK MOTIF | authKW | 116395 | 0% | 100% | 3 |
| 8 | NON AUG INITIATION | authKW | 107769 | 1% | 56% | 5 |
| 9 | UPSTREAM OPEN READING FRAME UORF | authKW | 96991 | 1% | 50% | 5 |
| 10 | RIBO SEQ | authKW | 88678 | 1% | 57% | 4 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Biochemistry & Molecular Biology | 4233 | 60% | 0% | 472 |
| 2 | Cell Biology | 1276 | 22% | 0% | 172 |
| 3 | Genetics & Heredity | 1075 | 16% | 0% | 129 |
| 4 | Biotechnology & Applied Microbiology | 1013 | 16% | 0% | 128 |
| 5 | Biochemical Research Methods | 622 | 10% | 0% | 80 |
| 6 | Mathematical & Computational Biology | 430 | 5% | 0% | 36 |
| 7 | Plant Sciences | 293 | 10% | 0% | 78 |
| 8 | Biophysics | 160 | 6% | 0% | 49 |
| 9 | Biology | 103 | 4% | 0% | 31 |
| 10 | Virology | 78 | 3% | 0% | 23 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | COLUMBIA UNIV MD PHD PROGRAM | 77597 | 0% | 100% | 2 |
| 2 | BACTERIAL INFECTSHINJUKU KU | 38798 | 0% | 100% | 1 |
| 3 | BELGIUM CFP | 38798 | 0% | 100% | 1 |
| 4 | BIOBIX MATH MODELLING STAT BIOINFORMAT | 38798 | 0% | 100% | 1 |
| 5 | BIOCHEM BOELELAAN 1083 | 38798 | 0% | 100% | 1 |
| 6 | BIOENGN BIOTECHNOL STATE ETHN AFFAIRS C | 38798 | 0% | 100% | 1 |
| 7 | CBIIGBMCCNRSUMR 7104U964 | 38798 | 0% | 100% | 1 |
| 8 | COLUMBIA SULZBERGER GENOME | 38798 | 0% | 100% | 1 |
| 9 | DIPARTIMENTO FIS BIOCHIM GEN | 38798 | 0% | 100% | 1 |
| 10 | DIRECT SCI VIVANTCOMMISSARIAT ENERGIA ATOM | 38798 | 0% | 100% | 1 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | NUCLEIC ACIDS RESEARCH | 4214 | 8% | 0% | 66 |
| 2 | MOLECULAR SYSTEMS BIOLOGY | 4139 | 1% | 1% | 9 |
| 3 | PLANT BIOTECHNOLOGY | 2786 | 1% | 1% | 7 |
| 4 | GENOME RESEARCH | 2269 | 1% | 1% | 9 |
| 5 | MOLECULAR AND CELLULAR BIOLOGY | 1742 | 4% | 0% | 32 |
| 6 | WILEY INTERDISCIPLINARY REVIEWS-RNA | 1684 | 1% | 1% | 4 |
| 7 | RNA | 1513 | 1% | 0% | 10 |
| 8 | ELIFE | 1301 | 1% | 0% | 10 |
| 9 | MOLECULAR & CELLULAR PROTEOMICS | 1242 | 1% | 0% | 10 |
| 10 | GENOME BIOLOGY | 1038 | 1% | 0% | 9 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | RIBOSOME PROFILING | 496181 | 3% | 47% | 27 | Search RIBOSOME+PROFILING | Search RIBOSOME+PROFILING |
| 2 | UORF | 405536 | 4% | 37% | 28 | Search UORF | Search UORF |
| 3 | UPSTREAM OPEN READING FRAME | 225715 | 2% | 36% | 16 | Search UPSTREAM+OPEN+READING+FRAME | Search UPSTREAM+OPEN+READING+FRAME |
| 4 | SCANNING MODEL | 190107 | 1% | 70% | 7 | Search SCANNING+MODEL | Search SCANNING+MODEL |
| 5 | AUG CONTEXT | 174589 | 1% | 75% | 6 | Search AUG+CONTEXT | Search AUG+CONTEXT |
| 6 | ARABIDOPSIS THALIANA T87 | 116395 | 0% | 100% | 3 | Search ARABIDOPSIS+THALIANA+T87 | Search ARABIDOPSIS+THALIANA+T87 |
| 7 | KOZAK MOTIF | 116395 | 0% | 100% | 3 | Search KOZAK+MOTIF | Search KOZAK+MOTIF |
| 8 | NON AUG INITIATION | 107769 | 1% | 56% | 5 | Search NON+AUG+INITIATION | Search NON+AUG+INITIATION |
| 9 | UPSTREAM OPEN READING FRAME UORF | 96991 | 1% | 50% | 5 | Search UPSTREAM+OPEN+READING+FRAME+UORF | Search UPSTREAM+OPEN+READING+FRAME+UORF |
| 10 | RIBO SEQ | 88678 | 1% | 57% | 4 | Search RIBO+SEQ | Search RIBO+SEQ |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | ANDREWS, SJ , ROTHNAGEL, JA , (2014) EMERGING EVIDENCE FOR FUNCTIONAL PEPTIDES ENCODED BY SHORT OPEN READING FRAMES.NATURE REVIEWS GENETICS. VOL. 15. ISSUE 3. P. 193-204 | 63 | 55% | 94 |
| 2 | HELLENS, RP , BROWN, CM , CHISNAL, MAW , WATERHOUSE, PM , MACKNIGHT, RC , (2016) THE EMERGING WORLD OF SMALL ORFS.TRENDS IN PLANT SCIENCE. VOL. 21. ISSUE 4. P. 317 -328 | 46 | 53% | 5 |
| 3 | BRAR, GA , WEISSMAN, JS , (2015) RIBOSOME PROFILING REVEALS THE WHAT, WHEN, WHERE AND HOW OF PROTEIN SYNTHESIS.NATURE REVIEWS MOLECULAR CELL BIOLOGY. VOL. 16. ISSUE 11. P. 651 -664 | 45 | 42% | 21 |
| 4 | PUEYO, JI , MAGNY, EG , COUSO, JP , (2016) NEW PEPTIDES UNDER THE S(ORF)ACE OF THE GENOME.TRENDS IN BIOCHEMICAL SCIENCES. VOL. 41. ISSUE 8. P. 665 -678 | 44 | 57% | 0 |
| 5 | MOUILLERON, H , DELCOURT, V , ROUCOU, X , (2016) DEATH OF A DOGMA: EUKARYOTIC MRNAS CAN CODE FOR MORE THAN ONE PROTEIN.NUCLEIC ACIDS RESEARCH. VOL. 44. ISSUE 1. P. 14 -23 | 50 | 41% | 2 |
| 6 | TAKAHASHI, H , TAKAHASHI, A , NAITO, S , ONOUCHI, H , (2012) BAIUCAS: A NOVEL BLAST-BASED ALGORITHM FOR THE IDENTIFICATION OF UPSTREAM OPEN READING FRAMES WITH CONSERVED AMINO ACID SEQUENCES AND ITS APPLICATION TO THE ARABIDOPSIS THALIANA GENOME.BIOINFORMATICS. VOL. 28. ISSUE 17. P. 2231-2241 | 27 | 84% | 13 |
| 7 | INGOLIA, NT , (2016) RIBOSOME FOOTPRINT PROFILING OF TRANSLATION THROUGHOUT THE GENOME.CELL. VOL. 165. ISSUE 1. P. 22 -33 | 35 | 38% | 10 |
| 8 | WETHMAR, K , (2014) THE REGULATORY POTENTIAL OF UPSTREAM OPEN READING FRAMES IN EUKARYOTIC GENE EXPRESSION.WILEY INTERDISCIPLINARY REVIEWS-RNA. VOL. 5. ISSUE 6. P. 765 -778 | 44 | 43% | 22 |
| 9 | JOHNSTONE, TG , BAZZINI, AA , GIRALDEZ, AJ , (2016) UPSTREAM ORFS ARE PREVALENT TRANSLATIONAL REPRESSORS IN VERTEBRATES.EMBO JOURNAL. VOL. 35. ISSUE 7. P. 706 -723 | 35 | 40% | 8 |
| 10 | MICHEL, AM , ANDREEV, DE , BARANOV, PV , (2014) COMPUTATIONAL APPROACH FOR CALCULATING THE PROBABILITY OF EUKARYOTIC TRANSLATION INITIATION FROM RIBO-SEQ DATA THAT TAKES INTO ACCOUNT LEAKY SCANNING.BMC BIOINFORMATICS. VOL. 15. ISSUE . P. - | 25 | 83% | 7 |
Classes with closest relation at Level 1 |