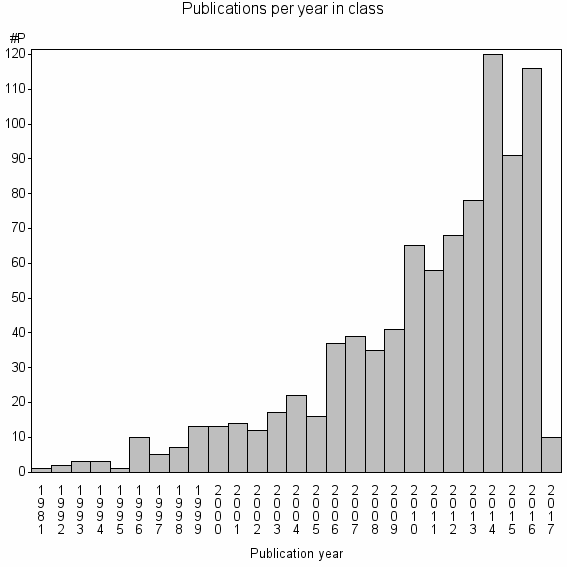
Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
| 12560 | 897 | 59.3 | 92% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 3 | 4 | PLANT SCIENCES//AGRONOMY//FOOD SCIENCE & TECHNOLOGY | 1882812 |
| 7 | 3 | PLANT SCIENCES//PLANT PHYSIOLOGY//ARABIDOPSIS | 149859 |
| 88 | 2 | PLANT SCIENCES//SALICYLIC ACID//JASMONIC ACID | 26878 |
| 12560 | 1 | RECEPTOR LIKE KINASE//FLG22//RLK | 897 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | RECEPTOR LIKE KINASE | authKW | 369797 | 4% | 29% | 37 |
| 2 | FLG22 | authKW | 307193 | 2% | 48% | 19 |
| 3 | RLK | authKW | 215206 | 2% | 45% | 14 |
| 4 | PLANT IMMUNITY | authKW | 213518 | 4% | 16% | 40 |
| 5 | WALL ASSOCIATED KINASE | authKW | 212091 | 1% | 69% | 9 |
| 6 | RECEPTOR LIKE PROTEIN KINASE | authKW | 205933 | 1% | 55% | 11 |
| 7 | LECTIN RECEPTOR LIKE KINASE | authKW | 204241 | 1% | 100% | 6 |
| 8 | LRR RLK | authKW | 189103 | 1% | 56% | 10 |
| 9 | MAMPS | authKW | 185559 | 1% | 42% | 13 |
| 10 | LECTIN RECEPTOR KINASE | authKW | 185327 | 1% | 78% | 7 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Plant Sciences | 20578 | 66% | 0% | 589 |
| 2 | Biochemistry & Molecular Biology | 1742 | 38% | 0% | 343 |
| 3 | Cell Biology | 777 | 17% | 0% | 149 |
| 4 | Parasitiology | 296 | 4% | 0% | 34 |
| 5 | Biotechnology & Applied Microbiology | 260 | 9% | 0% | 77 |
| 6 | Virology | 167 | 4% | 0% | 34 |
| 7 | Genetics & Heredity | 115 | 6% | 0% | 55 |
| 8 | Microbiology | 113 | 6% | 0% | 55 |
| 9 | Biochemical Research Methods | 33 | 3% | 0% | 27 |
| 10 | Agronomy | 23 | 2% | 0% | 18 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | ZURICH BASEL PLANT SCI | 104305 | 4% | 10% | 32 |
| 2 | ZMBP PLANT BIOCHEM | 90771 | 0% | 67% | 4 |
| 3 | PLANT BIOCHEM ZMBP | 68080 | 0% | 100% | 2 |
| 4 | PLANT MOL BIOL ZMBP PLANT BIOCHEM | 68080 | 0% | 100% | 2 |
| 5 | PLANT MICROBE INTERACT | 65519 | 3% | 7% | 28 |
| 6 | PLANT GENOM BIOTECHNOL | 49647 | 2% | 8% | 18 |
| 7 | SAINSBURY | 34053 | 3% | 4% | 24 |
| 8 | ABT PROTEOM METAB FORSHUNG 7 | 34040 | 0% | 100% | 1 |
| 9 | ANALYT ZMBP | 34040 | 0% | 100% | 1 |
| 10 | B353 LEVINE SCI | 34040 | 0% | 100% | 1 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | CURRENT OPINION IN PLANT BIOLOGY | 14111 | 3% | 2% | 27 |
| 2 | MOLECULAR PLANT PATHOLOGY | 13802 | 2% | 2% | 22 |
| 3 | MOLECULAR PLANT-MICROBE INTERACTIONS | 12221 | 4% | 1% | 35 |
| 4 | PLANT CELL | 10027 | 5% | 1% | 42 |
| 5 | PLANT SIGNALING & BEHAVIOR | 9580 | 1% | 3% | 11 |
| 6 | PLANT JOURNAL | 8263 | 5% | 1% | 42 |
| 7 | MOLECULAR PLANT | 6922 | 2% | 1% | 14 |
| 8 | FRONTIERS IN PLANT SCIENCE | 6575 | 3% | 1% | 31 |
| 9 | CELL HOST & MICROBE | 6208 | 2% | 1% | 14 |
| 10 | PLANT PHYSIOLOGY | 5957 | 7% | 0% | 61 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | RECEPTOR LIKE KINASE | 369797 | 4% | 29% | 37 | Search RECEPTOR+LIKE+KINASE | Search RECEPTOR+LIKE+KINASE |
| 2 | FLG22 | 307193 | 2% | 48% | 19 | Search FLG22 | Search FLG22 |
| 3 | RLK | 215206 | 2% | 45% | 14 | Search RLK | Search RLK |
| 4 | PLANT IMMUNITY | 213518 | 4% | 16% | 40 | Search PLANT+IMMUNITY | Search PLANT+IMMUNITY |
| 5 | WALL ASSOCIATED KINASE | 212091 | 1% | 69% | 9 | Search WALL+ASSOCIATED+KINASE | Search WALL+ASSOCIATED+KINASE |
| 6 | RECEPTOR LIKE PROTEIN KINASE | 205933 | 1% | 55% | 11 | Search RECEPTOR+LIKE+PROTEIN+KINASE | Search RECEPTOR+LIKE+PROTEIN+KINASE |
| 7 | LECTIN RECEPTOR LIKE KINASE | 204241 | 1% | 100% | 6 | Search LECTIN+RECEPTOR+LIKE+KINASE | Search LECTIN+RECEPTOR+LIKE+KINASE |
| 8 | LRR RLK | 189103 | 1% | 56% | 10 | Search LRR+RLK | Search LRR+RLK |
| 9 | MAMPS | 185559 | 1% | 42% | 13 | Search MAMPS | Search MAMPS |
| 10 | LECTIN RECEPTOR KINASE | 185327 | 1% | 78% | 7 | Search LECTIN+RECEPTOR+KINASE | Search LECTIN+RECEPTOR+KINASE |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | MACHO, AP , ZIPFEL, C , (2014) PLANT PRRS AND THE ACTIVATION OF INNATE IMMUNE SIGNALING.MOLECULAR CELL. VOL. 54. ISSUE 2. P. 263 -272 | 76 | 82% | 138 |
| 2 | ZIPFEL, C , (2014) PLANT PATTERN-RECOGNITION RECEPTORS.TRENDS IN IMMUNOLOGY. VOL. 35. ISSUE 7. P. 345 -351 | 59 | 63% | 140 |
| 3 | TRDA, L , BOUTROT, F , CLAVERIE, J , BRULE, D , DOREY, S , POINSSOT, B , (2015) PERCEPTION OF PATHOGENIC OR BENEFICIAL BACTERIA AND THEIR EVASION OF HOST IMMUNITY: PATTERN RECOGNITION RECEPTORS IN THE FRONTLINE.FRONTIERS IN PLANT SCIENCE. VOL. 6. ISSUE . P. - | 86 | 63% | 11 |
| 4 | COUTO, D , ZIPFEL, C , (2016) REGULATION OF PATTERN RECOGNITION RECEPTOR SIGNALLING IN PLANTS.NATURE REVIEWS IMMUNOLOGY. VOL. 16. ISSUE 9. P. 537 -552 | 97 | 48% | 5 |
| 5 | LI, L , YU, YF , ZHOU, ZY , ZHOU, JM , (2016) PLANT PATTERN-RECOGNITION RECEPTORS CONTROLLING INNATE IMMUNITY.SCIENCE CHINA-LIFE SCIENCES. VOL. 59. ISSUE 9. P. 878 -888 | 83 | 66% | 0 |
| 6 | YAMADA, K , YAMASHITA-YAMADA, M , HIRASE, T , FUJIWARA, T , TSUDA, K , HIRUMA, K , SAIJO, Y , (2016) DANGER PEPTIDE RECEPTOR SIGNALING IN PLANTS ENSURES BASAL IMMUNITY UPON PATHOGEN-INDUCED DEPLETION OF BAK1.EMBO JOURNAL. VOL. 35. ISSUE 1. P. 46 -61 | 51 | 59% | 12 |
| 7 | BOHM, H , ALBERT, I , FAN, L , REINHARD, A , NURNBERGER, T , (2014) IMMUNE RECEPTOR COMPLEXES AT THE PLANT CELL SURFACE.CURRENT OPINION IN PLANT BIOLOGY. VOL. 20. ISSUE . P. 47 -54 | 48 | 75% | 52 |
| 8 | MONAGHAN, J , ZIPFEL, C , (2012) PLANT PATTERN RECOGNITION RECEPTOR COMPLEXES AT THE PLASMA MEMBRANE.CURRENT OPINION IN PLANT BIOLOGY. VOL. 15. ISSUE 4. P. 349 -357 | 45 | 65% | 172 |
| 9 | HOLTON, N , NEKRASOV, V , RONALD, PC , ZIPFEL, C , (2015) THE PHYLOGENETICALLY-RELATED PATTERN RECOGNITION RECEPTORS EFR AND XA21 RECRUIT SIMILAR IMMUNE SIGNALING COMPONENTS IN MONOCOTS AND DICOTS.PLOS PATHOGENS. VOL. 11. ISSUE 1. P. - | 51 | 74% | 10 |
| 10 | HELFT, L , THOMPSON, M , BENT, AF , (2016) DIRECTED EVOLUTION OF FLS2 TOWARDS NOVEL FLAGELLIN PEPTIDE RECOGNITION.PLOS ONE. VOL. 11. ISSUE 6. P. - | 62 | 67% | 0 |
Classes with closest relation at Level 1 |