Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
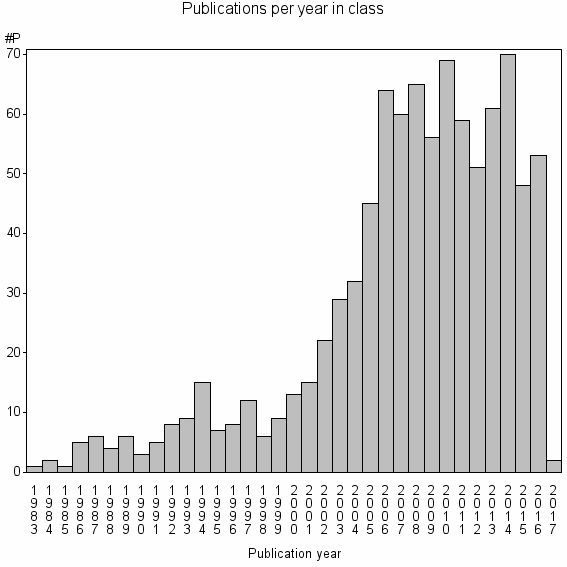
| 12242 | 921 | 31.1 | 74% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 90 | 3 | MATHEMATICAL & COMPUTATIONAL BIOLOGY//BIOINFORMATICS//BMC BIOINFORMATICS | 77178 |
| 3201 | 2 | GRAPHICAL REPRESENTATION//AUTHORSHIP ATTRIBUTION//CODE CLONE | 2089 |
| 12242 | 1 | GRAPHICAL REPRESENTATION//ALIGNMENT FREE//ALIGNMENT FREE METHOD | 921 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | GRAPHICAL REPRESENTATION | authKW | 1146577 | 8% | 46% | 76 |
| 2 | ALIGNMENT FREE | authKW | 575373 | 3% | 54% | 32 |
| 3 | ALIGNMENT FREE METHOD | authKW | 350175 | 1% | 81% | 13 |
| 4 | NUMERICAL CHARACTERIZATION | authKW | 270738 | 2% | 58% | 14 |
| 5 | SIMILARITIES DISSIMILARITIES | authKW | 265225 | 1% | 100% | 8 |
| 6 | CHAOS GAME REPRESENTATION | authKW | 238681 | 2% | 40% | 18 |
| 7 | ALIGNMENT FREE SEQUENCE COMPARISON | authKW | 235753 | 1% | 89% | 8 |
| 8 | PROTEOMICS MAPS | authKW | 232072 | 1% | 100% | 7 |
| 9 | LZ COMPLEXITY | authKW | 180497 | 1% | 78% | 7 |
| 10 | T LIFE | address | 176066 | 2% | 30% | 18 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Mathematical & Computational Biology | 21391 | 28% | 0% | 258 |
| 2 | Computer Science, Interdisciplinary Applications | 4305 | 19% | 0% | 173 |
| 3 | Mathematics, Interdisciplinary Applications | 3040 | 13% | 0% | 123 |
| 4 | Biochemical Research Methods | 2334 | 17% | 0% | 160 |
| 5 | Biology | 1250 | 11% | 0% | 101 |
| 6 | Biotechnology & Applied Microbiology | 923 | 15% | 0% | 134 |
| 7 | Evolutionary Biology | 691 | 6% | 0% | 56 |
| 8 | Chemistry, Multidisciplinary | 469 | 17% | 0% | 157 |
| 9 | Physics, Atomic, Molecular & Chemical | 384 | 9% | 0% | 85 |
| 10 | Genetics & Heredity | 384 | 10% | 0% | 90 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | T LIFE | 176066 | 2% | 30% | 18 |
| 2 | PRACOWNIA CHEMOMETRII SRODOWISKA | 66306 | 0% | 100% | 2 |
| 3 | SHANGHAI SYST BIOL | 66306 | 0% | 100% | 2 |
| 4 | COMP COMMUN | 48281 | 5% | 3% | 47 |
| 5 | RADIOL INFORMAT STAT | 45114 | 1% | 19% | 7 |
| 6 | ACCADEMIA NAZL LINCEI DEI | 33153 | 0% | 100% | 1 |
| 7 | AGR HYDRAUL ENGN | 33153 | 0% | 100% | 1 |
| 8 | AIZU CLUSTER MED ENGN INFORMAT ARC MED | 33153 | 0% | 100% | 1 |
| 9 | ALGORITHMS BIOINFORMAT COMPLEX FORMAL METHODS | 33153 | 0% | 100% | 1 |
| 10 | BETH ISRAEL DEACONESS MED GENOM | 33153 | 0% | 100% | 1 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | MATCH-COMMUNICATIONS IN MATHEMATICAL AND IN COMPUTER CHEMISTRY | 77497 | 6% | 4% | 59 |
| 2 | JOURNAL OF MATHEMATICAL CHEMISTRY | 10683 | 3% | 1% | 29 |
| 3 | JOURNAL OF THEORETICAL BIOLOGY | 9923 | 6% | 1% | 57 |
| 4 | ALGORITHMS FOR MOLECULAR BIOLOGY | 9472 | 1% | 3% | 9 |
| 5 | JOURNAL OF COMPUTATIONAL BIOLOGY | 9349 | 2% | 1% | 21 |
| 6 | SAR AND QSAR IN ENVIRONMENTAL RESEARCH | 8229 | 2% | 2% | 14 |
| 7 | EVOLUTIONARY BIOINFORMATICS | 6720 | 1% | 3% | 8 |
| 8 | CURRENT BIOINFORMATICS | 4785 | 1% | 2% | 8 |
| 9 | CURRENT COMPUTER-AIDED DRUG DESIGN | 4750 | 1% | 2% | 7 |
| 10 | JOURNAL OF COMPUTATIONAL AND THEORETICAL NANOSCIENCE | 4563 | 2% | 1% | 21 |
Author Key Words |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | BIELINSKA-WAZ, D , (2011) GRAPHICAL AND NUMERICAL REPRESENTATIONS OF DNA SEQUENCES: STATISTICAL ASPECTS OF SIMILARITY.JOURNAL OF MATHEMATICAL CHEMISTRY. VOL. 49. ISSUE 10. P. 2345 -2407 | 96 | 76% | 20 |
| 2 | YAO, YH , KONG, F , DAI, Q , HE, PA , (2013) A SEQUENCE-SEGMENTED METHOD APPLIED TO THE SIMILARITY ANALYSIS OF LONG PROTEIN SEQUENCE.MATCH-COMMUNICATIONS IN MATHEMATICAL AND IN COMPUTER CHEMISTRY. VOL. 70. ISSUE 1. P. 431-450 | 59 | 92% | 4 |
| 3 | QI, ZH , JIN, MZ , WANG, JS , LI, SL , (2015) NOVEL DNA SEQUENCE COMPARISON METHOD BASED ON MARKOV CHAIN AND INFORMATION ENTROPY.MINI-REVIEWS IN ORGANIC CHEMISTRY. VOL. 12. ISSUE 6. P. 524 -533 | 54 | 93% | 1 |
| 4 | RANDIC, M , NOVIC, M , PLAVSIC, D , (2013) MILESTONES IN GRAPHICAL BIOINFORMATICS.INTERNATIONAL JOURNAL OF QUANTUM CHEMISTRY. VOL. 113. ISSUE 22. P. 2413-2446 | 72 | 62% | 16 |
| 5 | RANDIC, M , ZUPAN, J , BALABAN, AT , VIKIC-TOPIC, D , PLAVSIC, D , (2011) GRAPHICAL REPRESENTATION OF PROTEINS.CHEMICAL REVIEWS. VOL. 111. ISSUE 2. P. 790 -862 | 82 | 51% | 61 |
| 6 | RANDIC, M , NOVIC, M , CHOUDHURY, AR , PLAVSIC, D , (2012) ON GRAPHICAL REPRESENTATION OF TRANS-MEMBRANE PROTEINS.SAR AND QSAR IN ENVIRONMENTAL RESEARCH. VOL. 23. ISSUE 3-4. P. 327-343 | 47 | 100% | 9 |
| 7 | YAO, YH , YAN, SJ , HAN, JN , DAI, Q , HE, PA , (2014) A NOVEL DESCRIPTOR OF PROTEIN SEQUENCES AND ITS APPLICATION.JOURNAL OF THEORETICAL BIOLOGY. VOL. 347. ISSUE . P. 109-117 | 49 | 82% | 14 |
| 8 | QI, ZH , JIN, MZ , YANG, H , (2015) A MEASURE OF PROTEIN SEQUENCE CHARACTERISTICS BASED ON THE FREQUENCY AND THE POSITION ENTROPY OF EXISTING K-WORDS.MATCH-COMMUNICATIONS IN MATHEMATICAL AND IN COMPUTER CHEMISTRY. VOL. 73. ISSUE 3. P. 731 -748 | 46 | 96% | 0 |
| 9 | YU, HJ , HUANG, DS , (2012) NOVEL GRAPHICAL REPRESENTATION OF GENOME SEQUENCE AND ITS APPLICATIONS IN SIMILARITY ANALYSIS.PHYSICA A-STATISTICAL MECHANICS AND ITS APPLICATIONS. VOL. 391. ISSUE 23. P. 6128-6136 | 47 | 94% | 5 |
| 10 | NANDY, A , HARLE, M , BASAK, SC , (2006) MATHEMATICAL DESCRIPTORS OF DNA SEQUENCES: DEVELOPMENT AND APPLICATIONS.ARKIVOC. VOL. . ISSUE . P. 211 -238 | 50 | 91% | 82 |
Classes with closest relation at Level 1 |