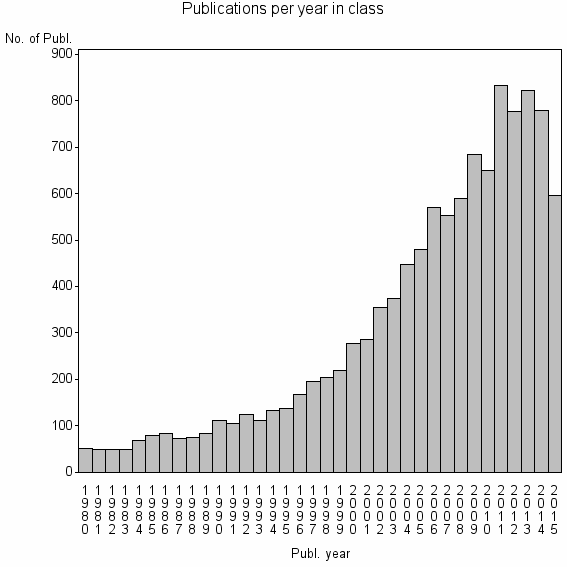
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 825 | 11237 | 42.4 | 72% |
Classes in level above (level 3) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 362 | 30818 | JOURNAL OF CHEMICAL INFORMATION AND COMPUTER SCIENCES//MATCH-COMMUNICATIONS IN MATHEMATICAL AND IN COMPUTER CHEMISTRY//JOURNAL OF CHEMICAL INFORMATION AND MODELING |
Classes in level below (level 1) |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | JOURNAL OF CHEMICAL INFORMATION AND MODELING | Journal | 628 | 42% | 10% | 1161 |
| 2 | B IT | Address | 336 | 85% | 2% | 180 |
| 3 | JOURNAL OF CHEMICAL INFORMATION AND COMPUTER SCIENCES | Journal | 331 | 31% | 8% | 882 |
| 4 | LIMES PROGRAM UNIT CHEM BIOL MED CHEM | Address | 297 | 87% | 1% | 144 |
| 5 | LIFE SCI INFORMAT | Address | 274 | 71% | 2% | 221 |
| 6 | JOURNAL OF CHEMINFORMATICS | Journal | 202 | 68% | 2% | 177 |
| 7 | VIRTUAL SCREENING | Author keyword | 172 | 29% | 5% | 511 |
| 8 | JOURNAL OF COMPUTER-AIDED MOLECULAR DESIGN | Journal | 139 | 27% | 4% | 442 |
| 9 | MOLECULAR SIMILARITY | Author keyword | 75 | 48% | 1% | 114 |
| 10 | CHEMICAL SPACE | Author keyword | 61 | 61% | 1% | 65 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | VIRTUAL SCREENING | 172 | 29% | 5% | 511 | Search VIRTUAL+SCREENING | Search VIRTUAL+SCREENING |
| 2 | MOLECULAR SIMILARITY | 75 | 48% | 1% | 114 | Search MOLECULAR+SIMILARITY | Search MOLECULAR+SIMILARITY |
| 3 | CHEMICAL SPACE | 61 | 61% | 1% | 65 | Search CHEMICAL+SPACE | Search CHEMICAL+SPACE |
| 4 | LIBRARY DESIGN | 60 | 60% | 1% | 65 | Search LIBRARY+DESIGN | Search LIBRARY+DESIGN |
| 5 | SIMILARITY SEARCHING | 53 | 59% | 1% | 59 | Search SIMILARITY+SEARCHING | Search SIMILARITY+SEARCHING |
| 6 | CHEMOGENOMICS | 50 | 52% | 1% | 67 | Search CHEMOGENOMICS | Search CHEMOGENOMICS |
| 7 | CHEMINFORMATICS | 48 | 43% | 1% | 85 | Search CHEMINFORMATICS | Search CHEMINFORMATICS |
| 8 | POLYPHARMACOLOGY | 42 | 52% | 1% | 58 | Search POLYPHARMACOLOGY | Search POLYPHARMACOLOGY |
| 9 | CHEMOINFORMATICS | 40 | 36% | 1% | 89 | Search CHEMOINFORMATICS | Search CHEMOINFORMATICS |
| 10 | SCORING FUNCTION | 40 | 40% | 1% | 78 | Search SCORING+FUNCTION | Search SCORING+FUNCTION |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | FLEXIBLE DOCKING | 391 | 60% | 4% | 432 |
| 2 | DRUG DISCOVERY | 299 | 15% | 16% | 1793 |
| 3 | MOLECULAR DOCKING | 226 | 36% | 4% | 505 |
| 4 | LEAD DISCOVERY | 222 | 59% | 2% | 250 |
| 5 | AUTOMATED DOCKING | 156 | 35% | 3% | 364 |
| 6 | SCORING FUNCTIONS | 151 | 47% | 2% | 236 |
| 7 | HIGH THROUGHPUT DOCKING | 141 | 70% | 1% | 116 |
| 8 | PROTEIN LIGAND DOCKING | 137 | 61% | 1% | 147 |
| 9 | LEAD GENERATION | 133 | 68% | 1% | 117 |
| 10 | PDBBIND DATABASE | 125 | 75% | 1% | 91 |
Journals |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | JOURNAL OF CHEMICAL INFORMATION AND MODELING | 628 | 42% | 10% | 1161 |
| 2 | JOURNAL OF CHEMICAL INFORMATION AND COMPUTER SCIENCES | 331 | 31% | 8% | 882 |
| 3 | JOURNAL OF CHEMINFORMATICS | 202 | 68% | 2% | 177 |
| 4 | JOURNAL OF COMPUTER-AIDED MOLECULAR DESIGN | 139 | 27% | 4% | 442 |
| 5 | MOLECULAR INFORMATICS | 57 | 34% | 1% | 137 |
| 6 | DRUG DISCOVERY TODAY | 35 | 12% | 2% | 266 |
| 7 | COMBINATORIAL CHEMISTRY & HIGH THROUGHPUT SCREENING | 13 | 10% | 1% | 125 |
| 8 | CURRENT COMPUTER-AIDED DRUG DESIGN | 12 | 19% | 0% | 55 |
| 9 | PERSPECTIVES IN DRUG DISCOVERY AND DESIGN | 10 | 25% | 0% | 36 |
| 10 | CURRENT OPINION IN DRUG DISCOVERY & DEVELOPMENT | 10 | 12% | 1% | 76 |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Natural Products As Sources of New Drugs over the 30 Years from 1981 to 2010 | 2012 | 904 | 105 | 32% |
| Natural products as sources of new drugs over the last 25 years | 2007 | 1738 | 69 | 32% |
| The Chemical Space Project | 2015 | 4 | 82 | 88% |
| Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings | 2001 | 2793 | 40 | 33% |
| The French National Compound Library: advances and future prospects | 2015 | 3 | 6 | 83% |
| InChI - the worldwide chemical structure identifier standard | 2013 | 49 | 2 | 100% |
| Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings | 1997 | 3504 | 40 | 33% |
| Network pharmacology: the next paradigm in drug discovery | 2008 | 679 | 96 | 43% |
| Evolutions in fragment-based drug design: the deconstruction-reconstruction approach | 2015 | 3 | 55 | 67% |
| A decade of fragment-based drug design: strategic advances and lessons learned | 2007 | 466 | 67 | 64% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | B IT | 336 | 85% | 1.6% | 180 |
| 2 | LIMES PROGRAM UNIT CHEM BIOL MED CHEM | 297 | 87% | 1.3% | 144 |
| 3 | LIFE SCI INFORMAT | 274 | 71% | 2.0% | 221 |
| 4 | UNILEVER MOL SCI INFORMAT | 42 | 42% | 0.7% | 78 |
| 5 | CHINESE PEDIAT | 30 | 100% | 0.1% | 12 |
| 6 | BIOINFORMAT ZBH | 24 | 91% | 0.1% | 10 |
| 7 | CHEMOGENOM | 21 | 71% | 0.2% | 17 |
| 8 | LIMES PROGRAM | 19 | 80% | 0.1% | 12 |
| 9 | DECS GLOBAL COMPOUND SCI | 17 | 100% | 0.1% | 8 |
| 10 | COMP AIDED DRUG DISCOVERY | 15 | 38% | 0.3% | 32 |
Related classes at same level (level 2) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000022608 | WIENER INDEX//SZEGED INDEX//QSAR |
| 2 | 0.0000017817 | BIOPARTITIONING MICELLAR CHROMATOGRAPHY//LIPOPHILICITY//IMMOBILIZED LIPOSOME CHROMATOGRAPHY |
| 3 | 0.0000014166 | HIV 1 PROTEASE//HIV PROTEASE//KNI 272 |
| 4 | 0.0000011561 | PROTEINS-STRUCTURE FUNCTION AND BIOINFORMATICS//PROTEIN STRUCTURE PREDICTION//PROTEIN FOLDING |
| 5 | 0.0000009928 | COMPUTER ASSISTED STRUCTURE ELUCIDATION//AUTOMATED CHEMISTRY WORKSTATION//SEARCH PREFILTERS |
| 6 | 0.0000009540 | LY573636//TASISULAM//CHEM TECHNOL DRUGS |
| 7 | 0.0000008834 | JOURNAL OF BIOMOLECULAR SCREENING//ANTIBODY MICROARRAYS//PROTEIN MICROARRAY |
| 8 | 0.0000008821 | PEPT1//PEPT2//HPEPT1 |
| 9 | 0.0000007922 | JOURNAL OF MOLECULAR GRAPHICS//HOPFIELD STYLE NETWORK//DRUG DOCKING |
| 10 | 0.0000006795 | PEPTIDE DEFORMYLASE//ENOYL ACP REDUCTASE//PLATENSIMYCIN |