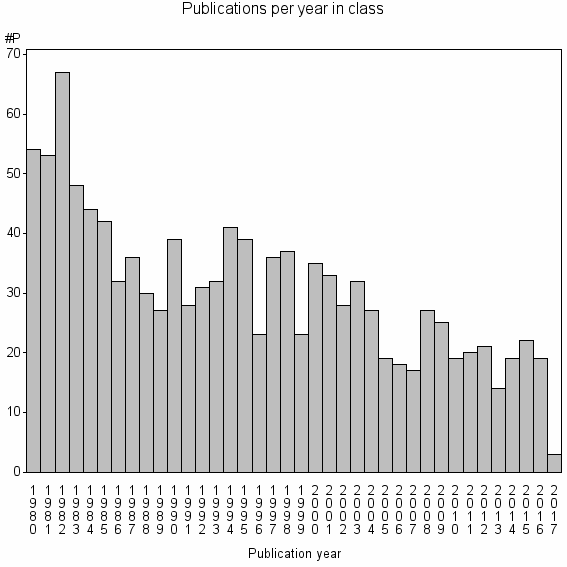
Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
| 9381 | 1160 | 39.5 | 65% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 71 | 3 | GENETICS & HEREDITY//CHROMATIN//CELL BIOLOGY | 83302 |
| 1094 | 2 | NUCLEAR MATRIX//LINKER HISTONE//HISTONE H1 | 9829 |
| 9381 | 1 | PROT SECT//HMG PROTEINS//HIGH MOBILITY GROUP PROTEINS | 1160 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | PROT SECT | address | 1019320 | 4% | 79% | 49 |
| 2 | HMG PROTEINS | authKW | 486426 | 3% | 60% | 31 |
| 3 | HIGH MOBILITY GROUP PROTEINS | authKW | 333361 | 3% | 33% | 38 |
| 4 | HIGH MOBILITY GROUP HMG PROTEINS | authKW | 247127 | 1% | 72% | 13 |
| 5 | HMG BOX | authKW | 207974 | 3% | 23% | 35 |
| 6 | HMGN2 | authKW | 188008 | 1% | 71% | 10 |
| 7 | HMGN5 | authKW | 184254 | 1% | 100% | 7 |
| 8 | HMGN | authKW | 164001 | 1% | 69% | 9 |
| 9 | NSBP1 | authKW | 157932 | 1% | 100% | 6 |
| 10 | HMG1 | authKW | 143291 | 1% | 39% | 14 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Biochemistry & Molecular Biology | 10007 | 74% | 0% | 864 |
| 2 | Biophysics | 2093 | 16% | 0% | 187 |
| 3 | Cell Biology | 1161 | 18% | 0% | 205 |
| 4 | Genetics & Heredity | 137 | 6% | 0% | 69 |
| 5 | Plant Sciences | 95 | 6% | 0% | 65 |
| 6 | CYTOLOGY & HISTOLOGY | 78 | 0% | 0% | 5 |
| 7 | Developmental Biology | 66 | 2% | 0% | 22 |
| 8 | Biology | 55 | 3% | 0% | 31 |
| 9 | Biochemical Research Methods | 35 | 3% | 0% | 33 |
| 10 | Oncology | 16 | 4% | 0% | 49 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | PROT SECT | 1019320 | 4% | 79% | 49 |
| 2 | ANAL CHROMOSOMAL PROT | 109673 | 0% | 83% | 5 |
| 3 | PROT SECT METAB | 78966 | 0% | 100% | 3 |
| 4 | FRONTIER COMP SCI | 35095 | 0% | 67% | 2 |
| 5 | UNIT INFECT IMMUN | 33018 | 1% | 16% | 8 |
| 6 | ANAL CHROMOSOMAL PROTEINS | 26322 | 0% | 100% | 1 |
| 7 | BIOL CELULAR MOLGRP EXPREIA | 26322 | 0% | 100% | 1 |
| 8 | BIOL POB 208103 | 26322 | 0% | 100% | 1 |
| 9 | BIOSCI RICE BIOCHEM CELL BIOL | 26322 | 0% | 100% | 1 |
| 10 | CELL BIOL GENE EXP S GRP RECEPTOR BIOL | 26322 | 0% | 100% | 1 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | BIOCHIMICA ET BIOPHYSICA ACTA-GENE REGULATORY MECHANISMS | 4900 | 1% | 1% | 14 |
| 2 | NUCLEIC ACIDS RESEARCH | 4721 | 7% | 0% | 85 |
| 3 | COMPREHENSIVE BIOCHEMISTRY | 4385 | 0% | 17% | 1 |
| 4 | BIOCHEMISTRY | 2540 | 6% | 0% | 73 |
| 5 | SEMINARS IN CELL BIOLOGY | 2445 | 0% | 5% | 2 |
| 6 | JOURNAL OF BIOLOGICAL CHEMISTRY | 1898 | 9% | 0% | 108 |
| 7 | PLANT MOLECULAR BIOLOGY | 1867 | 2% | 0% | 22 |
| 8 | JOURNAL OF MOLECULAR BIOLOGY | 1409 | 3% | 0% | 36 |
| 9 | BIOCHIMICA ET BIOPHYSICA ACTA | 1268 | 3% | 0% | 38 |
| 10 | SPECTROSCOPY-AN INTERNATIONAL JOURNAL | 1193 | 0% | 1% | 5 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | HMG PROTEINS | 486426 | 3% | 60% | 31 | Search HMG+PROTEINS | Search HMG+PROTEINS |
| 2 | HIGH MOBILITY GROUP PROTEINS | 333361 | 3% | 33% | 38 | Search HIGH+MOBILITY+GROUP+PROTEINS | Search HIGH+MOBILITY+GROUP+PROTEINS |
| 3 | HIGH MOBILITY GROUP HMG PROTEINS | 247127 | 1% | 72% | 13 | Search HIGH+MOBILITY+GROUP+HMG+PROTEINS | Search HIGH+MOBILITY+GROUP+HMG+PROTEINS |
| 4 | HMG BOX | 207974 | 3% | 23% | 35 | Search HMG+BOX | Search HMG+BOX |
| 5 | HMGN2 | 188008 | 1% | 71% | 10 | Search HMGN2 | Search HMGN2 |
| 6 | HMGN5 | 184254 | 1% | 100% | 7 | Search HMGN5 | Search HMGN5 |
| 7 | HMGN | 164001 | 1% | 69% | 9 | Search HMGN | Search HMGN |
| 8 | NSBP1 | 157932 | 1% | 100% | 6 | Search NSBP1 | Search NSBP1 |
| 9 | HMG1 | 143291 | 1% | 39% | 14 | Search HMG1 | Search HMG1 |
| 10 | HMG BOX DOMAIN | 131610 | 0% | 100% | 5 | Search HMG+BOX+DOMAIN | Search HMG+BOX+DOMAIN |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | BUSTIN, M , REEVES, R , (1996) HIGH-MOBILITY-GROUP CHROMOSOMAL PROTEINS: ARCHITECTURAL COMPONENTS THAT FACILITATE CHROMATIN FUNCTION.PROGRESS IN NUCLEIC ACID RESEARCH AND MOLECULAR BIOLOGY, VOL 54. VOL. 54. ISSUE . P. 35-100 | 109 | 43% | 565 |
| 2 | BUSTIN, M , LEHN, DA , LANDSMAN, D , (1990) STRUCTURAL FEATURES OF THE HMG CHROMOSOMAL-PROTEINS AND THEIR GENES.BIOCHIMICA ET BIOPHYSICA ACTA. VOL. 1049. ISSUE 3. P. 231-243 | 76 | 79% | 406 |
| 3 | ZHU, N , HANSEN, U , (2010) TRANSCRIPTIONAL REGULATION BY HMGN PROTEINS.BIOCHIMICA ET BIOPHYSICA ACTA-GENE REGULATORY MECHANISMS. VOL. 1799. ISSUE 1-2. P. 74-79 | 54 | 86% | 11 |
| 4 | GRASSER, KD , LAUNHOLT, D , GRASSER, M , (2007) HIGH MOBILITY GROUP PROTEINS OF THE PLANT HMGB FAMILY: DYNAMIC CHROMATIN MODULATORS.BIOCHIMICA ET BIOPHYSICA ACTA-GENE STRUCTURE AND EXPRESSION. VOL. 1769. ISSUE 5-6. P. 346-357 | 76 | 62% | 19 |
| 5 | GRASSER, KD , (2003) CHROMATIN-ASSOCIATED HMGA AND HMGB PROTEINS: VERSATILE CO-REGULATORS OF DNA-DEPENDENT PROCESSES.PLANT MOLECULAR BIOLOGY. VOL. 53. ISSUE 3. P. 281-295 | 64 | 69% | 48 |
| 6 | POSTNIKOV, Y , BUSTIN, M , (2010) REGULATION OF CHROMATIN STRUCTURE AND FUNCTION BY HMGN PROTEINS.BIOCHIMICA ET BIOPHYSICA ACTA-GENE REGULATORY MECHANISMS. VOL. 1799. ISSUE 1-2. P. 62-68 | 49 | 72% | 20 |
| 7 | GONZALEZ-ROMERO, R , EIRIN-LOPEZ, JM , AUSIO, J , (2015) EVOLUTION OF HIGH MOBILITY GROUP NUCLEOSOME-BINDING PROTEINS AND ITS IMPLICATIONS FOR VERTEBRATE CHROMATIN SPECIALIZATION.MOLECULAR BIOLOGY AND EVOLUTION. VOL. 32. ISSUE 1. P. 121 -131 | 47 | 64% | 4 |
| 8 | LEE, KB , THOMAS, JO , (2000) THE EFFECT OF THE ACIDIC TAIL ON THE DNA-BINDING PROPERTIES OF THE HMG1,2 CLASS OF PROTEINS: INSIGHTS FROM TAIL SWITCHING AND TAIL REMOVAL.JOURNAL OF MOLECULAR BIOLOGY. VOL. 304. ISSUE 2. P. 135-149 | 57 | 78% | 57 |
| 9 | THOMAS, JO , TRAVERS, AA , (2001) HMG1 AND 2, AND RELATED 'ARCHITECTURAL' DNA-BINDING PROTEINS.TRENDS IN BIOCHEMICAL SCIENCES. VOL. 26. ISSUE 3. P. 167-174 | 44 | 68% | 431 |
| 10 | GRASSER, M , LENTZ, A , LICHOTA, J , MERKLE, T , GRASSER, KD , (2006) THE ARABIDOPSIS GENOME ENCODES STRUCTURALLY AND FUNCTIONALLY DIVERSE HMGB-TYPE PROTEINS.JOURNAL OF MOLECULAR BIOLOGY. VOL. 358. ISSUE 3. P. 654-664 | 43 | 83% | 25 |
Classes with closest relation at Level 1 |