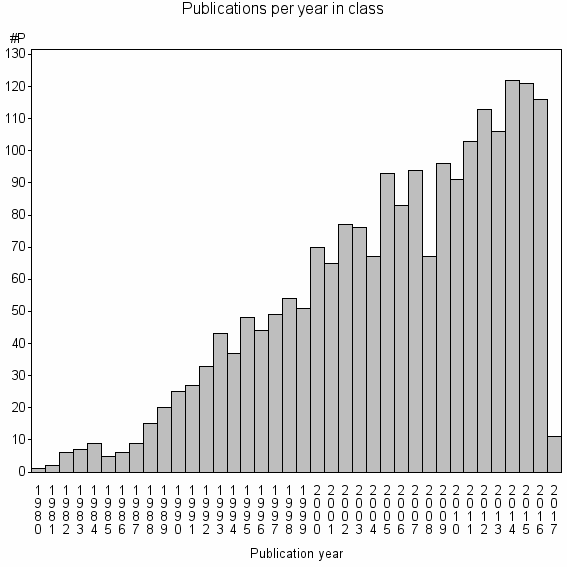
Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
| 3114 | 2062 | 52.0 | 83% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 3 | 4 | PLANT SCIENCES//AGRONOMY//FOOD SCIENCE & TECHNOLOGY | 1882812 |
| 40 | 3 | AGRONOMY//SOIL SCIENCE//AGRICULTURE, MULTIDISCIPLINARY | 100292 |
| 585 | 2 | NODULATION//FRANKIA//RHIZOBIUM | 14207 |
| 3114 | 1 | LOTUS JAPONICUS//SUPERNODULATION//MEDICAGO TRUNCATULA | 2062 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | LOTUS JAPONICUS | authKW | 865151 | 6% | 46% | 127 |
| 2 | SUPERNODULATION | authKW | 460683 | 2% | 84% | 37 |
| 3 | MEDICAGO TRUNCATULA | authKW | 445831 | 7% | 22% | 135 |
| 4 | NODULATION | authKW | 248558 | 7% | 12% | 144 |
| 5 | CARBOHYDRATE RECOGNIT SIGNALLING | address | 218431 | 1% | 49% | 30 |
| 6 | ARBUSCULAR MYCORRHIZA | authKW | 214332 | 8% | 9% | 156 |
| 7 | NODULATION MUTANTS | authKW | 208575 | 1% | 78% | 18 |
| 8 | SYMBIOSIS | authKW | 205930 | 12% | 6% | 238 |
| 9 | AUTOREGULATION OF NODULATION | authKW | 203764 | 1% | 81% | 17 |
| 10 | NOD FACTORS | authKW | 197127 | 2% | 28% | 48 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Plant Sciences | 61362 | 75% | 0% | 1537 |
| 2 | Biotechnology & Applied Microbiology | 1825 | 14% | 0% | 284 |
| 3 | Biochemistry & Molecular Biology | 1736 | 27% | 0% | 558 |
| 4 | Agronomy | 911 | 6% | 0% | 127 |
| 5 | Mycology | 680 | 2% | 0% | 51 |
| 6 | Soil Science | 549 | 3% | 0% | 70 |
| 7 | Cell Biology | 526 | 10% | 0% | 208 |
| 8 | Genetics & Heredity | 429 | 7% | 0% | 152 |
| 9 | Microbiology | 158 | 5% | 0% | 106 |
| 10 | Horticulture | 102 | 1% | 0% | 29 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | CARBOHYDRATE RECOGNIT SIGNALLING | 218431 | 1% | 49% | 30 |
| 2 | SYMBIOT SYST | 193672 | 1% | 59% | 22 |
| 3 | INTERACT PLANTES MICROORGANISMES | 116640 | 2% | 20% | 40 |
| 4 | LIPM | 100156 | 2% | 19% | 35 |
| 5 | GENET PLANT MICROBE INTERACT | 76148 | 0% | 86% | 6 |
| 6 | UMR441 | 70529 | 1% | 21% | 23 |
| 7 | PROGRAM MICROBIOL MOL BIOL BIOCHEM | 70498 | 0% | 48% | 10 |
| 8 | SCI VEGETAL | 67360 | 2% | 12% | 39 |
| 9 | EXCELLENCE INTEGRAT LEGUME | 64223 | 1% | 27% | 16 |
| 10 | UMR2594 | 62829 | 1% | 19% | 22 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | MOLECULAR PLANT-MICROBE INTERACTIONS | 157180 | 9% | 6% | 190 |
| 2 | PLANT PHYSIOLOGY | 27466 | 10% | 1% | 198 |
| 3 | NEW PHYTOLOGIST | 23832 | 6% | 1% | 123 |
| 4 | PLANT JOURNAL | 21633 | 5% | 1% | 103 |
| 5 | MYCORRHIZA | 14590 | 2% | 3% | 36 |
| 6 | PLANT CELL | 12428 | 3% | 1% | 71 |
| 7 | CURRENT OPINION IN PLANT BIOLOGY | 11498 | 2% | 2% | 37 |
| 8 | TRENDS IN PLANT SCIENCE | 7789 | 1% | 2% | 30 |
| 9 | PLANT AND CELL PHYSIOLOGY | 7629 | 3% | 1% | 60 |
| 10 | DNA RESEARCH | 6695 | 1% | 3% | 17 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | LOTUS JAPONICUS | 865151 | 6% | 46% | 127 | Search LOTUS+JAPONICUS | Search LOTUS+JAPONICUS |
| 2 | SUPERNODULATION | 460683 | 2% | 84% | 37 | Search SUPERNODULATION | Search SUPERNODULATION |
| 3 | MEDICAGO TRUNCATULA | 445831 | 7% | 22% | 135 | Search MEDICAGO+TRUNCATULA | Search MEDICAGO+TRUNCATULA |
| 4 | NODULATION | 248558 | 7% | 12% | 144 | Search NODULATION | Search NODULATION |
| 5 | ARBUSCULAR MYCORRHIZA | 214332 | 8% | 9% | 156 | Search ARBUSCULAR+MYCORRHIZA | Search ARBUSCULAR+MYCORRHIZA |
| 6 | NODULATION MUTANTS | 208575 | 1% | 78% | 18 | Search NODULATION+MUTANTS | Search NODULATION+MUTANTS |
| 7 | SYMBIOSIS | 205930 | 12% | 6% | 238 | Search SYMBIOSIS | Search SYMBIOSIS |
| 8 | AUTOREGULATION OF NODULATION | 203764 | 1% | 81% | 17 | Search AUTOREGULATION+OF+NODULATION | Search AUTOREGULATION+OF+NODULATION |
| 9 | NOD FACTORS | 197127 | 2% | 28% | 48 | Search NOD+FACTORS | Search NOD+FACTORS |
| 10 | ENOD40 | 170709 | 1% | 82% | 14 | Search ENOD40 | Search ENOD40 |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | OLDROYD, GED , (2013) SPEAK, FRIEND, AND ENTER: SIGNALLING SYSTEMS THAT PROMOTE BENEFICIAL SYMBIOTIC ASSOCIATIONS IN PLANTS.NATURE REVIEWS MICROBIOLOGY. VOL. 11. ISSUE 4. P. 252 -263 | 94 | 70% | 227 |
| 2 | OLDROYD, GED , DOWNIE, JM , (2008) COORDINATING NODULE MORPHOGENESIS WITH RHIZOBIAL INFECTION IN LEGUMES.ANNUAL REVIEW OF PLANT BIOLOGY. VOL. 59. ISSUE . P. 519-546 | 106 | 72% | 400 |
| 3 | PARNISKE, M , GUTJAHR, C , (2013) CELL AND DEVELOPMENTAL BIOLOGY OF ARBUSCULAR MYCORRHIZA SYMBIOSIS.ANNUAL REVIEW OF CELL AND DEVELOPMENTAL BIOLOGY, VOL 29. VOL. 29. ISSUE . P. 593 -617 | 106 | 70% | 81 |
| 4 | KOUCHI, H , IMAIZUMI-ANRAKU, H , HAYASHI, M , HAKOYAMA, T , NAKAGAWA, T , UMEHARA, Y , SUGANUMA, N , KAWAGUCHI, M , (2010) HOW MANY PEAS IN A POD? LEGUME GENES RESPONSIBLE FOR MUTUALISTIC SYMBIOSES UNDERGROUND.PLANT AND CELL PHYSIOLOGY. VOL. 51. ISSUE 9. P. 1381 -1397 | 110 | 83% | 69 |
| 5 | OLDROYD, GED , MURRAY, JD , POOLE, PS , DOWNIE, JA , (2011) THE RULES OF ENGAGEMENT IN THE LEGUME-RHIZOBIAL SYMBIOSIS.ANNUAL REVIEW OF GENETICS, VOL 45. VOL. 45. ISSUE . P. 119 -144 | 105 | 59% | 237 |
| 6 | JARDINAUD, MF , BOIVIN, S , RODDE, N , CATRICE, O , KISIALA, A , LEPAGE, A , MOREAU, S , ROUX, B , COTTRET, L , SALLET, E , ET AL (2016) A LASER DISSECTION-RNASEQ ANALYSIS HIGHLIGHTS THE ACTIVATION OF CYTOKININ PATHWAYS BY NOD FACTORS IN THE MEDICAGO TRUNCATULA ROOT EPIDERMIS.PLANT PHYSIOLOGY. VOL. 171. ISSUE 3. P. 2256 -2276 | 114 | 75% | 1 |
| 7 | MORTIER, V , HOLSTERS, M , GOORMACHTIG, S , (2012) NEVER TOO MANY? HOW LEGUMES CONTROL NODULE NUMBERS.PLANT CELL AND ENVIRONMENT. VOL. 35. ISSUE 2. P. 245 -258 | 117 | 70% | 42 |
| 8 | FERGUSON, BJ , MATHESIUS, U , (2014) PHYTOHORMONE REGULATION OF LEGUME-RHIZOBIA INTERACTIONS.JOURNAL OF CHEMICAL ECOLOGY. VOL. 40. ISSUE 7. P. 770 -790 | 117 | 66% | 16 |
| 9 | CAMPS, C , JARDINAUD, MF , RENGEL, D , CARRERE, S , HERVE, C , DEBELLE, F , GAMAS, P , BENSMIHEN, S , GOUGH, C , (2015) COMBINED GENETIC AND TRANSCRIPTOMIC ANALYSIS REVEALS THREE MAJOR SIGNALLING PATHWAYS ACTIVATED BY MYC-LCOS IN MEDICAGO TRUNCATULA.NEW PHYTOLOGIST. VOL. 208. ISSUE 1. P. 224 -240 | 90 | 84% | 7 |
| 10 | FERGUSON, BJ , INDRASUMUNAR, A , HAYASHI, S , LIN, MH , LIN, YH , REID, DE , GRESSHOFF, PM , (2010) MOLECULAR ANALYSIS OF LEGUME NODULE DEVELOPMENT AND AUTOREGULATION.JOURNAL OF INTEGRATIVE PLANT BIOLOGY. VOL. 52. ISSUE 1. P. 61 -76 | 81 | 76% | 142 |
Classes with closest relation at Level 1 |