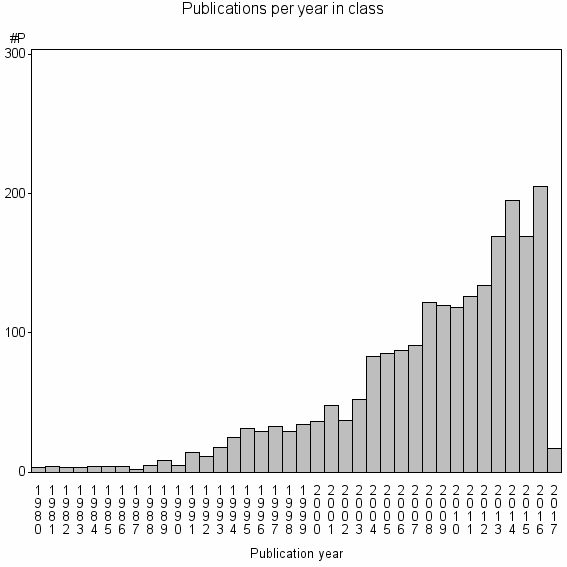
Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
| 2729 | 2163 | 55.0 | 81% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 469 | 3 | CHEMISTRY AND PHYSICS OF LIPIDS//BIOPHYSICAL JOURNAL//LIPID BILAYER | 24135 |
| 246 | 2 | LIPID BILAYER//CHEMISTRY AND PHYSICS OF LIPIDS//BIOPHYSICAL JOURNAL | 19868 |
| 2729 | 1 | MEMPHYS BIOMEMBRANE PHYS//LIPID BILAYER//BIOPHYS STAT MECH GRP | 2163 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | MEMPHYS BIOMEMBRANE PHYS | address | 205404 | 3% | 20% | 73 |
| 2 | LIPID BILAYER | authKW | 149210 | 6% | 9% | 121 |
| 3 | BIOPHYS STAT MECH GRP | address | 127014 | 1% | 43% | 21 |
| 4 | AREA PER LIPID | authKW | 121992 | 1% | 79% | 11 |
| 5 | PHYS CHEM DRUGS | address | 105107 | 1% | 24% | 31 |
| 6 | POPC | authKW | 102632 | 1% | 36% | 20 |
| 7 | CANADIAN NEUTRON BEAM | address | 97873 | 2% | 16% | 44 |
| 8 | BILAYER THICKNESS | authKW | 97645 | 1% | 43% | 16 |
| 9 | UIUC PROGRAM BIOPHYS | address | 70577 | 0% | 100% | 5 |
| 10 | GRP BIOINFORMAT MACROMOL BIOMAC | address | 69161 | 0% | 70% | 7 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Biophysics | 11398 | 27% | 0% | 577 |
| 2 | Chemistry, Physical | 10756 | 47% | 0% | 1017 |
| 3 | Physics, Atomic, Molecular & Chemical | 5088 | 20% | 0% | 431 |
| 4 | Biochemistry & Molecular Biology | 825 | 20% | 0% | 433 |
| 5 | Chemistry, Multidisciplinary | 667 | 14% | 0% | 304 |
| 6 | Computer Science, Interdisciplinary Applications | 287 | 4% | 0% | 81 |
| 7 | Mathematical & Computational Biology | 151 | 2% | 0% | 40 |
| 8 | Physics, Multidisciplinary | 125 | 5% | 0% | 114 |
| 9 | Materials Science, Multidisciplinary | 121 | 9% | 0% | 193 |
| 10 | Polymer Science | 110 | 4% | 0% | 85 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | MEMPHYS BIOMEMBRANE PHYS | 205404 | 3% | 20% | 73 |
| 2 | BIOPHYS STAT MECH GRP | 127014 | 1% | 43% | 21 |
| 3 | PHYS CHEM DRUGS | 105107 | 1% | 24% | 31 |
| 4 | CANADIAN NEUTRON BEAM | 97873 | 2% | 16% | 44 |
| 5 | UIUC PROGRAM BIOPHYS | 70577 | 0% | 100% | 5 |
| 6 | GRP BIOINFORMAT MACROMOL BIOMAC | 69161 | 0% | 70% | 7 |
| 7 | UIUC PROGRAM BIOENGN | 56461 | 0% | 100% | 4 |
| 8 | UIUC PROGRAM NEUROSCI | 56461 | 0% | 100% | 4 |
| 9 | COMPUTAT BIOPHYS BIOINFORMAT | 54197 | 1% | 30% | 13 |
| 10 | BIOMEMBRANE PHYS | 41742 | 1% | 14% | 21 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | BIOPHYSICAL JOURNAL | 43069 | 11% | 1% | 244 |
| 2 | JOURNAL OF PHYSICAL CHEMISTRY B | 35991 | 14% | 1% | 313 |
| 3 | JOURNAL OF CHEMICAL THEORY AND COMPUTATION | 30638 | 5% | 2% | 100 |
| 4 | BIOCHIMICA ET BIOPHYSICA ACTA-BIOMEMBRANES | 25810 | 5% | 2% | 107 |
| 5 | CHEMISTRY AND PHYSICS OF LIPIDS | 9999 | 2% | 1% | 48 |
| 6 | MOLECULAR SIMULATION | 5795 | 2% | 1% | 33 |
| 7 | JOURNAL OF COMPUTATIONAL CHEMISTRY | 5688 | 2% | 1% | 50 |
| 8 | SOFT MATTER | 5338 | 3% | 1% | 58 |
| 9 | JOURNAL OF CHEMICAL PHYSICS | 4094 | 7% | 0% | 162 |
| 10 | EUROPEAN BIOPHYSICS JOURNAL WITH BIOPHYSICS LETTERS | 3378 | 1% | 1% | 23 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | LIPID BILAYER | 149210 | 6% | 9% | 121 | Search LIPID+BILAYER | Search LIPID+BILAYER |
| 2 | AREA PER LIPID | 121992 | 1% | 79% | 11 | Search AREA+PER+LIPID | Search AREA+PER+LIPID |
| 3 | POPC | 102632 | 1% | 36% | 20 | Search POPC | Search POPC |
| 4 | BILAYER THICKNESS | 97645 | 1% | 43% | 16 | Search BILAYER+THICKNESS | Search BILAYER+THICKNESS |
| 5 | DMPC BILAYER | 45168 | 0% | 80% | 4 | Search DMPC+BILAYER | Search DMPC+BILAYER |
| 6 | POTENTIAL ENERGY PARAMETERS | 42346 | 0% | 100% | 3 | Search POTENTIAL+ENERGY+PARAMETERS | Search POTENTIAL+ENERGY+PARAMETERS |
| 7 | LATERAL PRESSURE PROFILE | 42340 | 0% | 50% | 6 | Search LATERAL+PRESSURE+PROFILE | Search LATERAL+PRESSURE+PROFILE |
| 8 | MOLECULAR DYNAMICS | 36890 | 10% | 1% | 210 | Search MOLECULAR+DYNAMICS | Search MOLECULAR+DYNAMICS |
| 9 | LIPID MEMBRANE | 31044 | 2% | 6% | 36 | Search LIPID+MEMBRANE | Search LIPID+MEMBRANE |
| 10 | MARTINI | 29401 | 0% | 42% | 5 | Search MARTINI | Search MARTINI |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | LYUBARTSEV, AP , RABINOVICH, AL , (2016) FORCE FIELD DEVELOPMENT FOR LIPID MEMBRANE SIMULATIONS.BIOCHIMICA ET BIOPHYSICA ACTA-BIOMEMBRANES. VOL. 1858. ISSUE 10. P. 2483 -2497 | 180 | 63% | 5 |
| 2 | LYUBARTSEV, AP , RABINOVICH, AL , (2011) RECENT DEVELOPMENT IN COMPUTER SIMULATIONS OF LIPID BILAYERS.SOFT MATTER. VOL. 7. ISSUE 1. P. 25 -39 | 172 | 67% | 81 |
| 3 | RABINOVICH, AL , LYUBARTSEV, AP , (2013) COMPUTER SIMULATION OF LIPID MEMBRANES: METHODOLOGY AND ACHIEVEMENTS.POLYMER SCIENCE SERIES C. VOL. 55. ISSUE 1. P. 162 -180 | 191 | 64% | 14 |
| 4 | ROG, T , VATTULAINEN, I , (2014) CHOLESTEROL, SPHINGOLIPIDS, AND GLYCOLIPIDS: WHAT DO WE KNOW ABOUT THEIR ROLE IN RAFT-LIKE MEMBRANES?.CHEMISTRY AND PHYSICS OF LIPIDS. VOL. 184. ISSUE . P. 82 -104 | 159 | 49% | 40 |
| 5 | POGER, D , CARON, B , MARK, AE , (2016) VALIDATING LIPID FORCE FIELDS AGAINST EXPERIMENTAL DATA: PROGRESS, CHALLENGES AND PERSPECTIVES.BIOCHIMICA ET BIOPHYSICA ACTA-BIOMEMBRANES. VOL. 1858. ISSUE 7. P. 1556 -1565 | 98 | 66% | 5 |
| 6 | OLLILA, OHS , PABST, G , (2016) ATOMISTIC RESOLUTION STRUCTURE AND DYNAMICS OF LIPID BILAYERS IN SIMULATIONS AND EXPERIMENTS.BIOCHIMICA ET BIOPHYSICA ACTA-BIOMEMBRANES. VOL. 1858. ISSUE 10. P. 2512 -2528 | 102 | 61% | 5 |
| 7 | AWOONOR-WILLIAMS, E , ROWLEY, CN , (2016) MOLECULAR SIMULATION OF NONFACILITATED MEMBRANE PERMEATION.BIOCHIMICA ET BIOPHYSICA ACTA-BIOMEMBRANES. VOL. 1858. ISSUE 7. P. 1672 -1687 | 89 | 50% | 8 |
| 8 | SHINODA, W , (2016) PERMEABILITY ACROSS LIPID MEMBRANES.BIOCHIMICA ET BIOPHYSICA ACTA-BIOMEMBRANES. VOL. 1858. ISSUE 10. P. 2254 -2265 | 92 | 63% | 2 |
| 9 | JAMBECK, JPM , LYUBARTSEV, AP , (2012) DERIVATION AND SYSTEMATIC VALIDATION OF A REFINED ALL-ATOM FORCE FIELD FOR PHOSPHATIDYLCHOLINE LIPIDS.JOURNAL OF PHYSICAL CHEMISTRY B. VOL. 116. ISSUE 10. P. 3164-3179 | 81 | 59% | 132 |
| 10 | NEALE, C , POMES, R , (2016) SAMPLING ERRORS IN FREE ENERGY SIMULATIONS OF SMALL MOLECULES IN LIPID BILAYERS.BIOCHIMICA ET BIOPHYSICA ACTA-BIOMEMBRANES. VOL. 1858. ISSUE 10. P. 2539 -2548 | 66 | 65% | 7 |
Classes with closest relation at Level 1 |