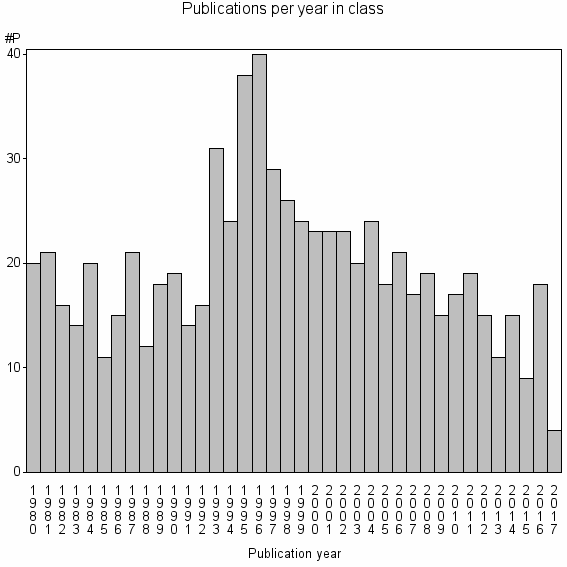
Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
| 14942 | 740 | 32.4 | 63% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | 3 ISOPROPYLMALATE DEHYDROGENASE | authKW | 1752859 | 6% | 90% | 47 |
| 2 | ISOCITRATE DEHYDROGENASE | authKW | 1474626 | 16% | 31% | 117 |
| 3 | ISOPROPYLMALATE DEHYDROGENASE | authKW | 647637 | 3% | 83% | 19 |
| 4 | HOMOISOCITRATE DEHYDROGENASE | authKW | 453889 | 1% | 100% | 11 |
| 5 | NADP ISOCITRATE DEHYDROGENASE | authKW | 311044 | 2% | 54% | 14 |
| 6 | BETA DECARBOXYLATING DEHYDROGENASE | authKW | 206313 | 1% | 100% | 5 |
| 7 | LEUB | authKW | 147364 | 1% | 71% | 5 |
| 8 | COENZYME SPECIFICITY | authKW | 130738 | 2% | 21% | 15 |
| 9 | ANCESTRAL RESIDUE | authKW | 123788 | 0% | 100% | 3 |
| 10 | COMMONOTE | authKW | 123788 | 0% | 100% | 3 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Biochemistry & Molecular Biology | 5794 | 71% | 0% | 527 |
| 2 | Biophysics | 1076 | 15% | 0% | 108 |
| 3 | Microbiology | 301 | 10% | 0% | 72 |
| 4 | Plant Sciences | 212 | 9% | 0% | 66 |
| 5 | Biochemical Research Methods | 142 | 6% | 0% | 41 |
| 6 | Biotechnology & Applied Microbiology | 135 | 7% | 0% | 53 |
| 7 | Food Science & Technology | 51 | 4% | 0% | 31 |
| 8 | Crystallography | 23 | 2% | 0% | 16 |
| 9 | Cell Biology | 20 | 5% | 0% | 36 |
| 10 | Chemistry, Applied | 15 | 2% | 0% | 16 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | TG BIOTECH CO LTD | 114614 | 1% | 56% | 5 |
| 2 | ANALYT BIOCHEM MICROBIOL | 82525 | 0% | 100% | 2 |
| 3 | ANALYT ULTRACENTRIFUGAT MACROMOL ASSEMBL | 41263 | 0% | 100% | 1 |
| 4 | BIOCHEM CELL BIOL EXCELLENCE MOL CELL | 41263 | 0% | 100% | 1 |
| 5 | BIOCHIM BIOPHYS GENET MICROORGANISMES BBGM | 41263 | 0% | 100% | 1 |
| 6 | BIOL VEGETAL MED AMBIENTE | 41263 | 0% | 100% | 1 |
| 7 | BIOTECHNOL PLANTES URA 1128 CNRS | 41263 | 0% | 100% | 1 |
| 8 | BIOTECMOL | 41263 | 0% | 100% | 1 |
| 9 | CNRS ERS569 | 41263 | 0% | 100% | 1 |
| 10 | CONSEJO SUPERIOR INVEST CIENTIF | 41263 | 0% | 100% | 1 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | PROTEIN ENGINEERING | 4648 | 2% | 1% | 16 |
| 2 | BIOCHEMISTRY | 3412 | 9% | 0% | 67 |
| 3 | ARCHIVES OF BIOCHEMISTRY AND BIOPHYSICS | 2603 | 4% | 0% | 33 |
| 4 | EXTREMOPHILES | 1854 | 1% | 1% | 8 |
| 5 | FREE RADICAL RESEARCH | 1663 | 1% | 0% | 11 |
| 6 | BIOCHIMIE | 1527 | 2% | 0% | 15 |
| 7 | JOURNAL OF BIOCHEMISTRY | 1509 | 3% | 0% | 20 |
| 8 | JOURNAL OF BIOLOGICAL CHEMISTRY | 1178 | 9% | 0% | 68 |
| 9 | ACTA CRYSTALLOGRAPHICA SECTION D-BIOLOGICAL CRYSTALLOGRAPHY | 1117 | 1% | 0% | 11 |
| 10 | BIOSCIENCE BIOTECHNOLOGY AND BIOCHEMISTRY | 853 | 2% | 0% | 16 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | 3 ISOPROPYLMALATE DEHYDROGENASE | 1752859 | 6% | 90% | 47 | Search 3+ISOPROPYLMALATE+DEHYDROGENASE | Search 3+ISOPROPYLMALATE+DEHYDROGENASE |
| 2 | ISOCITRATE DEHYDROGENASE | 1474626 | 16% | 31% | 117 | Search ISOCITRATE+DEHYDROGENASE | Search ISOCITRATE+DEHYDROGENASE |
| 3 | ISOPROPYLMALATE DEHYDROGENASE | 647637 | 3% | 83% | 19 | Search ISOPROPYLMALATE+DEHYDROGENASE | Search ISOPROPYLMALATE+DEHYDROGENASE |
| 4 | HOMOISOCITRATE DEHYDROGENASE | 453889 | 1% | 100% | 11 | Search HOMOISOCITRATE+DEHYDROGENASE | Search HOMOISOCITRATE+DEHYDROGENASE |
| 5 | NADP ISOCITRATE DEHYDROGENASE | 311044 | 2% | 54% | 14 | Search NADP+ISOCITRATE+DEHYDROGENASE | Search NADP+ISOCITRATE+DEHYDROGENASE |
| 6 | BETA DECARBOXYLATING DEHYDROGENASE | 206313 | 1% | 100% | 5 | Search BETA+DECARBOXYLATING+DEHYDROGENASE | Search BETA+DECARBOXYLATING+DEHYDROGENASE |
| 7 | LEUB | 147364 | 1% | 71% | 5 | Search LEUB | Search LEUB |
| 8 | COENZYME SPECIFICITY | 130738 | 2% | 21% | 15 | Search COENZYME+SPECIFICITY | Search COENZYME+SPECIFICITY |
| 9 | ANCESTRAL RESIDUE | 123788 | 0% | 100% | 3 | Search ANCESTRAL+RESIDUE | Search ANCESTRAL+RESIDUE |
| 10 | COMMONOTE | 123788 | 0% | 100% | 3 | Search COMMONOTE | Search COMMONOTE |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | MCALISTER-HENN, L , (2012) LIGAND BINDING AND STRUCTURAL CHANGES ASSOCIATED WITH ALLOSTERY IN YEAST NAD(+)-SPECIFIC ISOCITRATE DEHYDROGENASE.ARCHIVES OF BIOCHEMISTRY AND BIOPHYSICS. VOL. 519. ISSUE 2. P. 112 -117 | 38 | 90% | 6 |
| 2 | WANG, A , CAO, ZY , WANG, P , LIU, AM , PAN, W , WANG, J , ZHU, GP , (2011) HETEROEXPRESSION AND CHARACTERIZATION OF A MONOMERIC ISOCITRATE DEHYDROGENASE FROM THE MULTICELLULAR PROKARYOTE STREPTOMYCES AVERMITILIS MA-4680.MOLECULAR BIOLOGY REPORTS. VOL. 38. ISSUE 6. P. 3717-3724 | 35 | 85% | 2 |
| 3 | STEEN, IH , MADERN, D , KARLSTROM, M , LIEN, T , LADENSTEIN, R , BIRKELAND, NK , (2001) COMPARISON OF ISOCITRATE DEHYDROGENASE FROM THREE HYPERTHERMOPHILES REVEALS DIFFERENCES IN THERMOSTABILITY, COFACTOR SPECIFICITY, OLIGOMERIC STATE, AND PHYLOGENETIC AFFILIATION.JOURNAL OF BIOLOGICAL CHEMISTRY. VOL. 276. ISSUE 47. P. 43924 -43931 | 44 | 79% | 40 |
| 4 | STOKKE, R , MADERN, D , FEDOY, AE , KARLSEN, S , BIRKELAND, NK , STEEN, IH , (2007) BIOCHEMICAL CHARACTERIZATION OF ISOCITRATE DEHYDROGENASE FROM METHYLOCOCCUS CAPSULATUS REVEALS A UNIQUE NAD(+)-DEPENDENT HOMOTETRAMERIC ENZYME.ARCHIVES OF MICROBIOLOGY. VOL. 187. ISSUE 5. P. 361 -370 | 33 | 80% | 12 |
| 5 | NANGO, E , YAMAMOTO, T , KUMASAKA, T , EGUCHI, T , (2009) CRYSTAL STRUCTURE OF 3-ISOPROPYLMALATE DEHYDROGENASE IN COMPLEX WITH NAD(+) AND A DESIGNED INHIBITOR.BIOORGANIC & MEDICINAL CHEMISTRY. VOL. 17. ISSUE 22. P. 7789 -7794 | 30 | 81% | 6 |
| 6 | ZHANG, BB , WANG, BJ , WANG, P , CAO, ZY , HUANG, EQ , HAO, JS , DEAN, AM , ZHU, GP , (2009) ENZYMATIC CHARACTERIZATION OF A MONOMERIC ISOCITRATE DEHYDROGENASE FROM STREPTOMYCES LIVIDANS TK54.BIOCHIMIE. VOL. 91. ISSUE 11-12. P. 1405-1410 | 28 | 88% | 3 |
| 7 | SOUNDAR, S , O'HAGAN, M , FOMULU, KS , COLMAN, RF , (2006) IDENTIFICATION OF MN2+-BINDING ASPARTATES FROM ALPHA, BETA, AND GAMMA SUBUNITS OF HUMAN NAD-DEPENDENT ISOCITRATE DEHYDROGENASE.JOURNAL OF BIOLOGICAL CHEMISTRY. VOL. 281. ISSUE 30. P. 21073-21081 | 27 | 90% | 11 |
| 8 | WANG, P , SONG, P , JIN, MM , ZHU, GP , (2013) ISOCITRATE DEHYDROGENASE FROM STREPTOCOCCUS MUTANS: BIOCHEMICAL PROPERTIES AND EVALUATION OF A PUTATIVE PHOSPHORYLATION SITE AT SER102.PLOS ONE. VOL. 8. ISSUE 3. P. - | 31 | 65% | 2 |
| 9 | SIDHU, NS , DELBAERE, LTJ , SHELDRICK, GM , (2011) STRUCTURE OF A HIGHLY NADP(+)-SPECIFIC ISOCITRATE DEHYDROGENASE.ACTA CRYSTALLOGRAPHICA SECTION D-BIOLOGICAL CRYSTALLOGRAPHY. VOL. 67. ISSUE . P. 856 -869 | 37 | 56% | 3 |
| 10 | PENG, YJ , ZHONG, C , HUANG, W , DING, JP , (2008) STRUCTURAL STUDIES OF SACCHAROMYCES CEREVESIAE MITOCHONDRIAL NADP-DEPENDENT ISOCITRATE DEHYDROGENASE IN DIFFERENT ENZYMATIC STATES REVEAL SUBSTANTIAL CONFORMATIONAL CHANGES DURING THE CATALYTIC REACTION.PROTEIN SCIENCE. VOL. 17. ISSUE 9. P. 1542 -1554 | 32 | 67% | 18 |
Classes with closest relation at Level 1 |