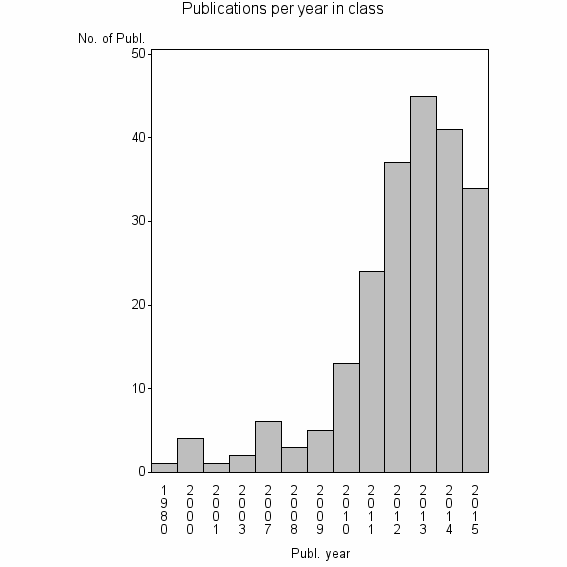
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 26161 | 216 | 41.0 | 83% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 1568 | 6715 | NEXT GENERATION SEQUENCING//RNA SEQ//COPY NUMBER VARIATION |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | MAPPING BY SEQUENCING | Author keyword | 6 | 80% | 2% | 4 |
| 2 | RADSEQ | Author keyword | 4 | 40% | 4% | 8 |
| 3 | REDUCED REPRESENTATION LIBRARIES | Author keyword | 3 | 100% | 1% | 3 |
| 4 | SHOREMAPPING | Author keyword | 3 | 100% | 1% | 3 |
| 5 | RAD SEQ | Author keyword | 3 | 26% | 4% | 9 |
| 6 | RAD TAG | Author keyword | 3 | 60% | 1% | 3 |
| 7 | REDUCED REPRESENTATION LIBRARY | Author keyword | 2 | 43% | 1% | 3 |
| 8 | GENOTYPING BY SEQUENCING | Author keyword | 1 | 13% | 5% | 10 |
| 9 | GENOTYPE BY SEQUENCING | Author keyword | 1 | 40% | 1% | 2 |
| 10 | REDUCED REPRESENTATION SEQUENCING | Author keyword | 1 | 40% | 1% | 2 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | MAPPING BY SEQUENCING | 6 | 80% | 2% | 4 | Search MAPPING+BY+SEQUENCING | Search MAPPING+BY+SEQUENCING |
| 2 | RADSEQ | 4 | 40% | 4% | 8 | Search RADSEQ | Search RADSEQ |
| 3 | REDUCED REPRESENTATION LIBRARIES | 3 | 100% | 1% | 3 | Search REDUCED+REPRESENTATION+LIBRARIES | Search REDUCED+REPRESENTATION+LIBRARIES |
| 4 | SHOREMAPPING | 3 | 100% | 1% | 3 | Search SHOREMAPPING | Search SHOREMAPPING |
| 5 | RAD SEQ | 3 | 26% | 4% | 9 | Search RAD+SEQ | Search RAD+SEQ |
| 6 | RAD TAG | 3 | 60% | 1% | 3 | Search RAD+TAG | Search RAD+TAG |
| 7 | REDUCED REPRESENTATION LIBRARY | 2 | 43% | 1% | 3 | Search REDUCED+REPRESENTATION+LIBRARY | Search REDUCED+REPRESENTATION+LIBRARY |
| 8 | GENOTYPING BY SEQUENCING | 1 | 13% | 5% | 10 | Search GENOTYPING+BY+SEQUENCING | Search GENOTYPING+BY+SEQUENCING |
| 9 | GENOTYPE BY SEQUENCING | 1 | 40% | 1% | 2 | Search GENOTYPE+BY+SEQUENCING | Search GENOTYPE+BY+SEQUENCING |
| 10 | REDUCED REPRESENTATION SEQUENCING | 1 | 40% | 1% | 2 | Search REDUCED+REPRESENTATION+SEQUENCING | Search REDUCED+REPRESENTATION+SEQUENCING |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | SNP DISCOVERY | 4 | 11% | 16% | 34 |
| 2 | MUTATION IDENTIFICATION | 4 | 56% | 2% | 5 |
| 3 | POLYMORPHISM DISCOVERY | 3 | 50% | 2% | 4 |
| 4 | RAD SEQ | 3 | 60% | 1% | 3 |
| 5 | REDUCED REPRESENTATION LIBRARY | 2 | 67% | 1% | 2 |
| 6 | REDUCED REPRESENTATION | 2 | 20% | 4% | 9 |
| 7 | RAD MARKERS | 2 | 18% | 5% | 10 |
| 8 | NUCLEOTIDE POLYMORPHISM DISCOVERY | 1 | 14% | 2% | 5 |
| 9 | ALFALFA GENOTYPES | 1 | 50% | 0% | 1 |
| 10 | CRATER LAKE CICHLIDS | 1 | 50% | 0% | 1 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Using next-generation sequencing to isolate mutant genes from forward genetic screens | 2014 | 13 | 111 | 33% |
| Genome-wide genetic marker discovery and genotyping using next-generation sequencing | 2011 | 445 | 73 | 22% |
| RAD in the realm of next-generation sequencing technologies | 2011 | 43 | 13 | 69% |
| Genotyping-by-Sequencing for Plant Breeding and Genetics | 2012 | 53 | 53 | 30% |
| Applications of next-generation sequencing to phylogeography and phylogenetics | 2013 | 76 | 117 | 14% |
| Getting started in mapping-by-sequencing | 2015 | 0 | 51 | 43% |
| Genotyping-by-sequencing (GBS), an ultimate marker-assisted selection (MAS) tool to accelerate plant breeding | 2014 | 7 | 71 | 17% |
| Perspective on Industrial Application of Specialized Metabolites in the Genomic Era | 2015 | 0 | 16 | 25% |
| Whole-genome resequencing: changing the paradigms of SNP detection, molecular mapping and gene discovery | 2015 | 0 | 76 | 18% |
| Diversification and asymmetrical gene flow across time and space: lineage sorting and hybridization in polytypic barking frogs | 2014 | 1 | 80 | 15% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | GENEPOOL | 1 | 23% | 1.4% | 3 |
| 2 | MONTPELLIER GENOMIX | 1 | 50% | 0.5% | 1 |
| 3 | OSEB UMR7205 | 1 | 50% | 0.5% | 1 |
| 4 | PANECO | 1 | 50% | 0.5% | 1 |
| 5 | UMR 5174ENFA | 1 | 50% | 0.5% | 1 |
| 6 | USDA ARS CROPS PATHOL GENET UNIT | 1 | 50% | 0.5% | 1 |
| 7 | ZOOTEHN BIOTEHNOL | 1 | 50% | 0.5% | 1 |
| 8 | GENEPOOL GENOM IL | 1 | 20% | 1.4% | 3 |
| 9 | SECT VERTEBRATE DEV | 0 | 33% | 0.5% | 1 |
| 10 | UAR 1209 | 0 | 33% | 0.5% | 1 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000209785 | GENOME ASSEMBLY//VARIANT CALLING//TARGET ENRICHMENT |
| 2 | 0.0000181601 | TILLING//SURVEYOR NUCLEASE//ECOTILLING |
| 3 | 0.0000122189 | FISH MOL GENET BIOTECHNOL//PROGRAM CELL MOL BIOSCI//AQUAT GENOM UNIT |
| 4 | 0.0000096110 | BLOOMINGTON DROSOPHILA STOCK//ENDS OUT TARGETING//ENDS OUT |
| 5 | 0.0000093583 | MCDONALD KREITMAN TEST//COALESCENT//SELECTIVE SWEEPS |
| 6 | 0.0000090295 | LONG GLUME//TRITICUM POLONICUM//TRITICUM PETROPAVLOVSKYI |
| 7 | 0.0000085344 | UNIVARIATE OPTIMIZATION//UNIDIMENSIONAL SEARCH METHOD//INITIAL INTERVAL OF UNCERTAINTY |
| 8 | 0.0000075860 | RNA SEQ//DE NOVO ASSEMBLY//TRANSCRIPTOME ASSEMBLY |
| 9 | 0.0000075521 | PHYLOGENETIC NETWORK//PHYLOGENETIC NETWORKS//TIGHT SPAN |
| 10 | 0.0000072054 | GASTEROSTEUS ACULEATUS//GASTEROSTEUS//STICKLEBACK |