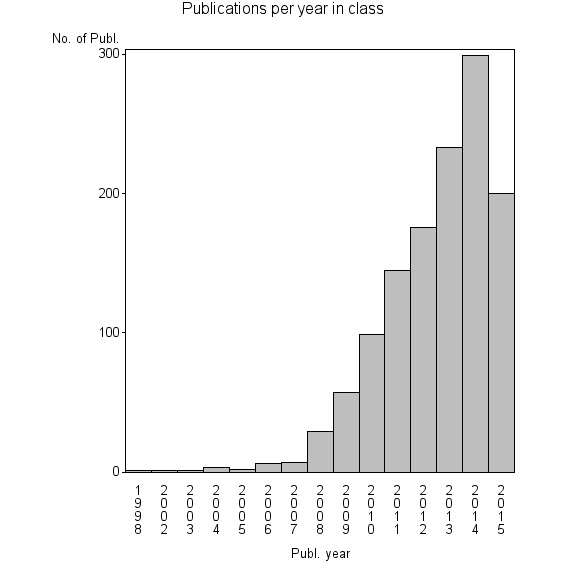
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 7698 | 1259 | 41.6 | 86% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 1568 | 6715 | NEXT GENERATION SEQUENCING//RNA SEQ//COPY NUMBER VARIATION |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | RNA SEQ | Author keyword | 37 | 16% | 17% | 218 |
| 2 | DE NOVO ASSEMBLY | Author keyword | 9 | 24% | 3% | 33 |
| 3 | TRANSCRIPTOME ASSEMBLY | Author keyword | 8 | 45% | 1% | 13 |
| 4 | ILLUMINA PAIRED END SEQUENCING | Author keyword | 5 | 60% | 0% | 6 |
| 5 | RNA SEQUENCING | Author keyword | 5 | 12% | 3% | 39 |
| 6 | TRANSCRIPT DISCOVERY | Author keyword | 4 | 75% | 0% | 3 |
| 7 | NSW SYST BIOL INITIAT | Address | 2 | 44% | 0% | 4 |
| 8 | ABI SOLID | Author keyword | 2 | 67% | 0% | 2 |
| 9 | CHAIR BIOINFORMAT GRP | Address | 2 | 67% | 0% | 2 |
| 10 | CLIN GENET BIOINFORMAT | Address | 2 | 67% | 0% | 2 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | RNA SEQ | 37 | 16% | 17% | 218 | Search RNA+SEQ | Search RNA+SEQ |
| 2 | DE NOVO ASSEMBLY | 9 | 24% | 3% | 33 | Search DE+NOVO+ASSEMBLY | Search DE+NOVO+ASSEMBLY |
| 3 | TRANSCRIPTOME ASSEMBLY | 8 | 45% | 1% | 13 | Search TRANSCRIPTOME+ASSEMBLY | Search TRANSCRIPTOME+ASSEMBLY |
| 4 | ILLUMINA PAIRED END SEQUENCING | 5 | 60% | 0% | 6 | Search ILLUMINA+PAIRED+END+SEQUENCING | Search ILLUMINA+PAIRED+END+SEQUENCING |
| 5 | RNA SEQUENCING | 5 | 12% | 3% | 39 | Search RNA+SEQUENCING | Search RNA+SEQUENCING |
| 6 | TRANSCRIPT DISCOVERY | 4 | 75% | 0% | 3 | Search TRANSCRIPT+DISCOVERY | Search TRANSCRIPT+DISCOVERY |
| 7 | ABI SOLID | 2 | 67% | 0% | 2 | Search ABI+SOLID | Search ABI+SOLID |
| 8 | DIGITAL GENE EXPRESSION TAG PROFILING DGE | 2 | 67% | 0% | 2 | Search DIGITAL+GENE+EXPRESSION+TAG+PROFILING+DGE | Search DIGITAL+GENE+EXPRESSION+TAG+PROFILING+DGE |
| 9 | REPLANTING DISEASE | 2 | 67% | 0% | 2 | Search REPLANTING+DISEASE | Search REPLANTING+DISEASE |
| 10 | SPOTTED SEAL PHOCA LARGHA | 2 | 67% | 0% | 2 | Search SPOTTED+SEAL+PHOCA+LARGHA | Search SPOTTED+SEAL+PHOCA+LARGHA |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | SEQUENCE COUNT DATA | 50 | 63% | 4% | 50 |
| 2 | DIFFERENTIAL EXPRESSION ANALYSIS | 33 | 26% | 9% | 109 |
| 3 | RNA SEQ DATA | 25 | 21% | 8% | 107 |
| 4 | SEQ DATA | 18 | 30% | 4% | 49 |
| 5 | MAMMALIAN TRANSCRIPTOMES | 16 | 61% | 1% | 17 |
| 6 | SPLICE JUNCTIONS | 15 | 34% | 3% | 35 |
| 7 | CELL TRANSCRIPTOME | 15 | 73% | 1% | 11 |
| 8 | ROOT TRANSCRIPTOME | 11 | 52% | 1% | 15 |
| 9 | SEQ | 11 | 15% | 5% | 67 |
| 10 | REFERENCE GENOME | 9 | 21% | 3% | 39 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| RNA-Seq: a revolutionary tool for transcriptomics | 2009 | 2552 | 36 | 28% |
| Next-generation transcriptome assembly | 2011 | 285 | 66 | 45% |
| RNA sequencing: advances, challenges and opportunities | 2011 | 414 | 100 | 30% |
| Computational methods for transcriptome annotation and quantification using RNA-seq | 2011 | 242 | 75 | 47% |
| RNA-Seq: revelation of the messengers | 2013 | 29 | 13 | 77% |
| Transcriptomics in the RNA-seq era | 2013 | 52 | 69 | 42% |
| RNA-Seq Technology and Its Application in Fish Transcriptomics | 2014 | 11 | 117 | 31% |
| Library preparation methods for next-generation sequencing: Tone down the bias | 2014 | 22 | 42 | 17% |
| Microarrays, deep sequencing and the true measure of the transcriptome | 2011 | 156 | 45 | 36% |
| From RNA-seq reads to differential expression results | 2010 | 166 | 84 | 48% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | NSW SYST BIOL INITIAT | 2 | 44% | 0.3% | 4 |
| 2 | CHAIR BIOINFORMAT GRP | 2 | 67% | 0.2% | 2 |
| 3 | CLIN GENET BIOINFORMAT | 2 | 67% | 0.2% | 2 |
| 4 | COOPERAT INNOVAT CULTIVAT UTILIZAT NONWOOD | 2 | 67% | 0.2% | 2 |
| 5 | FUNCT GENOM TECHNOL TEAM | 2 | 67% | 0.2% | 2 |
| 6 | STATE FO T TREE GENET BREEDING | 2 | 67% | 0.2% | 2 |
| 7 | SYST PHARMACOL BIOMARKERS | 2 | 67% | 0.2% | 2 |
| 8 | OMICS SCI | 2 | 17% | 0.9% | 11 |
| 9 | PREVENT MED DIAG INNOVAT PROGRAM PMI | 2 | 43% | 0.2% | 3 |
| 10 | AGRILIFE GENOM SERV | 1 | 100% | 0.2% | 2 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000180168 | GENOME ASSEMBLY//VARIANT CALLING//TARGET ENRICHMENT |
| 2 | 0.0000148597 | DIGITAL PCR//DROPLET DIGITAL PCR//SINGLE CELL TRANSCRIPTOMICS |
| 3 | 0.0000109858 | SPLICED LEADER//TRANS SPLICING//CIRCULAR RNA |
| 4 | 0.0000107270 | SERIAL ANALYSIS OF GENE EXPRESSION//SAGE//GLGI |
| 5 | 0.0000086015 | CHIP SEQ//PEAK CALLING//DNASE SEQ |
| 6 | 0.0000084945 | LONG NONCODING RNAS//LNCRNA//LONG NON CODING RNA |
| 7 | 0.0000075860 | MAPPING BY SEQUENCING//RADSEQ//REDUCED REPRESENTATION LIBRARIES |
| 8 | 0.0000075452 | SR PROTEINS//SR PROTEIN//ALTERNATIVE SPLICING |
| 9 | 0.0000072939 | EQTL//GENETICAL GENOMICS//ALLELE SPECIFIC GENE EXPRESSION |
| 10 | 0.0000063382 | CIS MOTIF//ARABIDOPSIS T87 CELL LINE//GENE ATLAS |