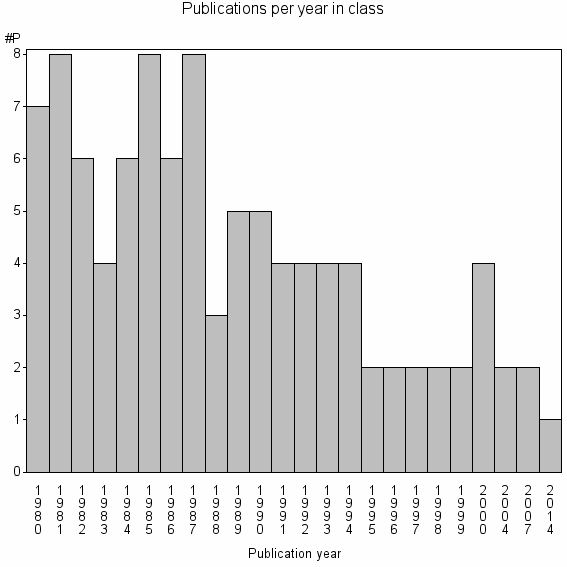
Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
| 35156 | 101 | 34.8 | 59% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 11 | 4 | NEUROSCIENCES//CLINICAL NEUROLOGY//NEUROL | 1112395 |
| 209 | 3 | GHRELIN//ADRENOMEDULLIN//NEUROPEPTIDE Y | 51926 |
| 1178 | 2 | CHOLECYSTOKININ//CCK B RECEPTOR//GASTRIN | 9326 |
| 35156 | 1 | AR42 J CELL//EUKARYOTIC GENE TRANSCRIPTION//MAMMALIAN REPORTER GENE | 101 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | AR42 J CELL | authKW | 302333 | 1% | 100% | 1 |
| 2 | EUKARYOTIC GENE TRANSCRIPTION | authKW | 302333 | 1% | 100% | 1 |
| 3 | MAMMALIAN REPORTER GENE | authKW | 302333 | 1% | 100% | 1 |
| 4 | MATURE EXPORT | authKW | 302333 | 1% | 100% | 1 |
| 5 | PANCREATIC 266 6 CELL | authKW | 302333 | 1% | 100% | 1 |
| 6 | PANCREATIC RIBONUCLEASE GENE | authKW | 302333 | 1% | 100% | 1 |
| 7 | RODENTIA ARVICOLINAE | authKW | 302333 | 1% | 100% | 1 |
| 8 | ELASTASE GENE | authKW | 151165 | 1% | 50% | 1 |
| 9 | TRANSCRIPTION ELEMENT | authKW | 151165 | 1% | 50% | 1 |
| 10 | DNAASE I FOOTPRINTING | authKW | 100776 | 1% | 33% | 1 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Biochemistry & Molecular Biology | 656 | 65% | 0% | 66 |
| 2 | Cell Biology | 236 | 26% | 0% | 26 |
| 3 | Genetics & Heredity | 209 | 20% | 0% | 20 |
| 4 | Developmental Biology | 32 | 4% | 0% | 4 |
| 5 | Evolutionary Biology | 30 | 4% | 0% | 4 |
| 6 | Biophysics | 6 | 4% | 0% | 4 |
| 7 | Biotechnology & Applied Microbiology | 3 | 4% | 0% | 4 |
| 8 | Biology | 2 | 2% | 0% | 2 |
| 9 | Endocrinology & Metabolism | 2 | 3% | 0% | 3 |
| 10 | Gastroenterology & Hepatology | 1 | 2% | 0% | 2 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | EMIL MANNKOPFF STR 2 | 75582 | 1% | 25% | 1 |
| 2 | BELGYOGYASZATI TANSZEK KLIN | 3641 | 1% | 1% | 1 |
| 3 | TROP MARINE SCI | 2136 | 2% | 0% | 2 |
| 4 | MOL BIOL ONCOL | 1924 | 1% | 1% | 1 |
| 5 | MED HUMAN GENET | 973 | 1% | 0% | 1 |
| 6 | CARDIOVASC SURG CLIN | 387 | 1% | 0% | 1 |
| 7 | VET POPULAT MED | 345 | 1% | 0% | 1 |
| 8 | MOL BIOL GENET | 306 | 3% | 0% | 3 |
| 9 | GEN BIOL | 301 | 1% | 0% | 1 |
| 10 | CHEM BIOENGN | 96 | 1% | 0% | 1 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | CRC CRITICAL REVIEWS IN BIOTECHNOLOGY | 3874 | 1% | 1% | 1 |
| 2 | UCLA SYMPOSIA ON MOLECULAR AND CELLULAR BIOLOGY | 2476 | 1% | 1% | 1 |
| 3 | BIOCHEMICAL GENETICS | 2176 | 4% | 0% | 4 |
| 4 | ISOZYMES-CURRENT TOPICS IN BIOLOGICAL AND MEDICAL RESEARCH | 2127 | 1% | 1% | 1 |
| 5 | MOLECULAR AND CELLULAR BIOLOGY | 1954 | 12% | 0% | 12 |
| 6 | FORTSCHRITTE DER ZOOLOGIE | 1285 | 1% | 0% | 1 |
| 7 | CARLSBERG RESEARCH COMMUNICATIONS | 890 | 1% | 0% | 1 |
| 8 | CANADIAN JOURNAL OF GASTROENTEROLOGY AND HEPATOLOGY | 741 | 1% | 0% | 1 |
| 9 | GENETIKA | 617 | 3% | 0% | 3 |
| 10 | CELL | 500 | 5% | 0% | 5 |
Author Key Words |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | AR42 J CELL | 302333 | 1% | 100% | 1 | Search AR42+J+CELL | Search AR42+J+CELL |
| 2 | EUKARYOTIC GENE TRANSCRIPTION | 302333 | 1% | 100% | 1 | Search EUKARYOTIC+GENE+TRANSCRIPTION | Search EUKARYOTIC+GENE+TRANSCRIPTION |
| 3 | MAMMALIAN REPORTER GENE | 302333 | 1% | 100% | 1 | Search MAMMALIAN+REPORTER+GENE | Search MAMMALIAN+REPORTER+GENE |
| 4 | MATURE EXPORT | 302333 | 1% | 100% | 1 | Search MATURE+EXPORT | Search MATURE+EXPORT |
| 5 | PANCREATIC 266 6 CELL | 302333 | 1% | 100% | 1 | Search PANCREATIC+266+6+CELL | Search PANCREATIC+266+6+CELL |
| 6 | PANCREATIC RIBONUCLEASE GENE | 302333 | 1% | 100% | 1 | Search PANCREATIC+RIBONUCLEASE+GENE | Search PANCREATIC+RIBONUCLEASE+GENE |
| 7 | RODENTIA ARVICOLINAE | 302333 | 1% | 100% | 1 | Search RODENTIA+ARVICOLINAE | Search RODENTIA+ARVICOLINAE |
| 8 | ELASTASE GENE | 151165 | 1% | 50% | 1 | Search ELASTASE+GENE | Search ELASTASE+GENE |
| 9 | TRANSCRIPTION ELEMENT | 151165 | 1% | 50% | 1 | Search TRANSCRIPTION+ELEMENT | Search TRANSCRIPTION+ELEMENT |
| 10 | DNAASE I FOOTPRINTING | 100776 | 1% | 33% | 1 | Search DNAASE+I+FOOTPRINTING | Search DNAASE+I+FOOTPRINTING |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | CRERAR, MM , ROOKS, NE , (1987) THE STRUCTURE AND EXPRESSION OF AMYLASE GENES IN MAMMALS - AN OVERVIEW.CRC CRITICAL REVIEWS IN BIOTECHNOLOGY. VOL. 5. ISSUE 3. P. 217-227 | 30 | 67% | 3 |
| 2 | MEISLER, MH , ANTONUCCI, TK , TREISMAN, LO , GUMUCIO, DL , SAMUELSON, LC , (1986) INTERSTRAIN VARIATION IN AMYLASE GENE COPY NUMBER AND MESSENGER-RNA ABUNDANCE IN 3 MOUSE-TISSUES.GENETICS. VOL. 113. ISSUE 3. P. 713-722 | 18 | 75% | 8 |
| 3 | KRUSE, F , ROSE, SD , SWIFT, GH , HAMMER, RE , MACDONALD, RJ , (1995) COOPERATION BETWEEN ELEMENTS OF AN ORGAN-SPECIFIC TRANSCRIPTIONAL ENHANCER IN ANIMALS.MOLECULAR AND CELLULAR BIOLOGY. VOL. 15. ISSUE 8. P. 4385-4394 | 16 | 47% | 23 |
| 4 | TOSI, M , BOVEY, R , ASTOLFI, S , BODARY, S , MEISLER, M , WELLAUER, PK , (1984) MULTIPLE NON-ALLELIC GENES ENCODING PANCREATIC ALPHA-AMYLASE OF MOUSE ARE EXPRESSED IN A STRAIN-SPECIFIC FASHION.EMBO JOURNAL. VOL. 3. ISSUE 12. P. 2809-2816 | 15 | 75% | 11 |
| 5 | COCKELL, M , STOLARCZYK, D , FRUTIGER, S , HUGHES, GJ , HAGENBUCHLE, O , WELLAUER, PK , (1995) BINDING-SITES FOR HEPATOCYTE NUCLEAR FACTOR-3-BETA OR FACTOR-3-GAMMA AND PANCREAS TRANSCRIPTION FACTOR-1 ARE REQUIRED FOR EFFICIENT EXPRESSION OF THE GENE ENCODING PANCREATIC ALPHA-AMYLASE.MOLECULAR AND CELLULAR BIOLOGY. VOL. 15. ISSUE 4. P. 1933-1941 | 14 | 42% | 45 |
| 6 | JONES, JM , KELLER, SA , SAMUELSON, LC , OSBORN, L , ROSENBERG, MP , MEISLER, MH , (1989) A SALIVARY AMYLASE TRANSGENE IS EFFICIENTLY EXPRESSED IN LIVER BUT NOT IN PAROTID-GLAND OF TRANSGENIC MICE.NUCLEIC ACIDS RESEARCH. VOL. 17. ISSUE 16. P. 6613-6623 | 15 | 56% | 5 |
| 7 | THOMSEN, KK , HJORTH, JP , NIELSEN, JT , (1984) GENETIC-VARIATION IN RESTRICTION PATTERNS AMONG MOUSE AMYLASE GENE COMPLEXES.BIOCHEMICAL GENETICS. VOL. 22. ISSUE 3-4. P. 257-273 | 13 | 87% | 4 |
| 8 | MEISLER, M , STRAHLER, J , WIEBAUER, K , THOMSEN, KK , (1983) MULTIPLE GENES ENCODE MOUSE PANCREATIC AMYLASES.ISOZYMES-CURRENT TOPICS IN BIOLOGICAL AND MEDICAL RESEARCH. VOL. 7. ISSUE . P. 39-57 | 13 | 87% | 3 |
| 9 | PAUL, PR , ELLIOTT, RW , (1987) ANALYSIS OF THE MOUSE AMY LOCUS IN RECOMBINANT INBRED MOUSE STRAINS.BIOCHEMICAL GENETICS. VOL. 25. ISSUE 7-8. P. 569-579 | 12 | 67% | 4 |
| 10 | NIELSEN, JT , (1982) VARIATION IN AMYLASE HAPLOTYPES AMONG CONGENIC LINES OF THE HOUSE MOUSE.GENETICS. VOL. 102. ISSUE 3. P. 571-582 | 10 | 100% | 8 |
Classes with closest relation at Level 1 |