Class information for: |
Basic class information |
| Class id | #P | Avg. number of references |
Database coverage of references |
|---|---|---|---|
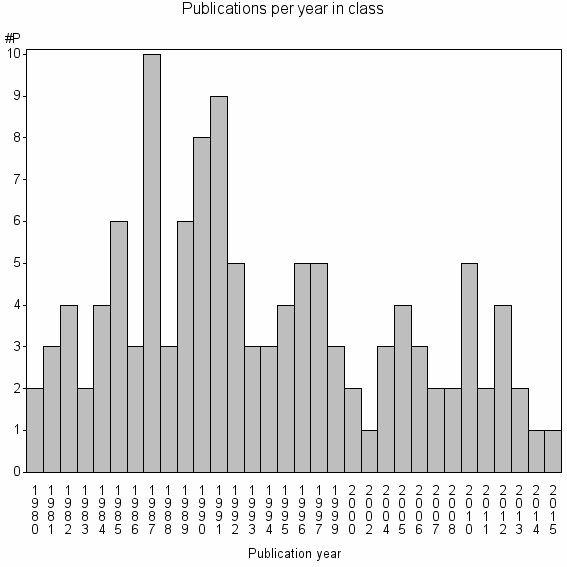
| 33744 | 120 | 28.3 | 47% |
Hierarchy of classes |
The table includes all classes above and classes immediately below the current class. |
| Cluster id | Level | Cluster label | #P |
|---|---|---|---|
| 0 | 4 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//ONCOLOGY | 4064930 |
| 469 | 3 | CHEMISTRY AND PHYSICS OF LIPIDS//BIOPHYSICAL JOURNAL//LIPID BILAYER | 24135 |
| 246 | 2 | LIPID BILAYER//CHEMISTRY AND PHYSICS OF LIPIDS//BIOPHYSICAL JOURNAL | 19868 |
| 33744 | 1 | ALTERNATING BILAYERS//ATOMIC MOLECULAR MODELS//DIPYRENYL PHOSPHOLIPIDS | 120 |
Terms with highest relevance score |
| rank | Term | termType | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|---|
| 1 | ALTERNATING BILAYERS | authKW | 254463 | 1% | 100% | 1 |
| 2 | ATOMIC MOLECULAR MODELS | authKW | 254463 | 1% | 100% | 1 |
| 3 | DIPYRENYL PHOSPHOLIPIDS | authKW | 254463 | 1% | 100% | 1 |
| 4 | FLUORESCENCE DISTRIBUTION ANALYSIS | authKW | 254463 | 1% | 100% | 1 |
| 5 | FLUORESCENCE PROBES RADIATIONLESS ENERGY TRANSFER | authKW | 254463 | 1% | 100% | 1 |
| 6 | LYSOZYME LIPOSOMES COMPLEXES | authKW | 254463 | 1% | 100% | 1 |
| 7 | MEMBRANE FREE VOLUME | authKW | 254463 | 1% | 100% | 1 |
| 8 | MIXED BINARY MONOLAYERS | authKW | 254463 | 1% | 100% | 1 |
| 9 | MODEL MEMBRANES STRUCTURE | authKW | 254463 | 1% | 100% | 1 |
| 10 | MONOLAYER MEMBRANE MODEL | authKW | 254463 | 1% | 100% | 1 |
Web of Science journal categories |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | Biophysics | 955 | 33% | 0% | 39 |
| 2 | Multidisciplinary Sciences | 340 | 9% | 0% | 11 |
| 3 | Chemistry, Physical | 84 | 20% | 0% | 24 |
| 4 | Biochemistry & Molecular Biology | 76 | 24% | 0% | 29 |
| 5 | Materials Science, Biomaterials | 68 | 4% | 0% | 5 |
| 6 | CYTOLOGY & HISTOLOGY | 32 | 1% | 0% | 1 |
| 7 | Materials Science, Coatings & Films | 19 | 3% | 0% | 4 |
| 8 | Cell Biology | 14 | 8% | 0% | 9 |
| 9 | Materials Science, Multidisciplinary | 13 | 11% | 0% | 13 |
| 10 | Biology | 10 | 3% | 0% | 4 |
Address terms |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | MOL BASIS NEURODEGENERAT CIMN | 145405 | 2% | 29% | 2 |
| 2 | FOOD SCI BURNSIDES | 127231 | 1% | 50% | 1 |
| 3 | BIOL MED PHYS | 120524 | 5% | 8% | 6 |
| 4 | DEP QUIM BIOQUIM | 114503 | 3% | 15% | 3 |
| 5 | ECOL SAFETY | 31802 | 3% | 4% | 3 |
| 6 | HELSINKI BIOPHYS BIOMEMBRANE GRP | 30604 | 3% | 3% | 4 |
| 7 | FARMA | 19572 | 1% | 8% | 1 |
| 8 | UMR6251 | 19572 | 1% | 8% | 1 |
| 9 | QUIM FIS MOL | 9956 | 5% | 1% | 6 |
| 10 | ELE SCI | 7950 | 1% | 3% | 1 |
Journals |
| Rank | Term | Chi square | Shr. of publ. in class containing term |
Class's shr. of term's tot. occurrences |
#P with term in class |
|---|---|---|---|---|---|
| 1 | MATERIALS SCIENCE & ENGINEERING C-BIOMIMETIC MATERIALS SENSORS AND SYSTEMS | 28272 | 1% | 11% | 1 |
| 2 | BIOFIZIKA | 13710 | 13% | 0% | 15 |
| 3 | BIOLOGICHESKIE MEMBRANY | 2381 | 4% | 0% | 5 |
| 4 | NUOVO CIMENTO DELLA SOCIETA ITALIANA DI FISICA C-GEOPHYSICS AND SPACE PHYSICS | 1698 | 2% | 0% | 2 |
| 5 | ZEITSCHRIFT FUR METEOROLOGIE | 1114 | 1% | 0% | 1 |
| 6 | BIOCHIMICA ET BIOPHYSICA ACTA-BIOMEMBRANES | 1014 | 4% | 0% | 5 |
| 7 | UKRAINSKII BIOKHIMICHESKII ZHURNAL | 879 | 2% | 0% | 2 |
| 8 | TEORETICHESKAYA I EKSPERIMENTALNAYA KHIMIYA | 873 | 2% | 0% | 2 |
| 9 | DOKLADY AKADEMII NAUK SSSR | 746 | 7% | 0% | 8 |
| 10 | DOKLADI NA BOLGARSKATA AKADEMIYA NA NAUKITE | 737 | 3% | 0% | 3 |
Author Key Words |
Core articles |
The table includes core articles in the class. The following variables is taken into account for the relevance score of an article in a cluster c: (1) Number of references referring to publications in the class. (2) Share of total number of active references referring to publications in the class. (3) Age of the article. New articles get higher score than old articles. (4) Citation rate, normalized to year. |
| Rank | Reference | # ref. in cl. |
Shr. of ref. in cl. |
Citations |
|---|---|---|---|---|
| 1 | AL KAYAL, T , NAPPINI, S , RUSSO, E , BERTI, D , BUCCIANTINI, M , STEFANI, M , BAGLIONI, P , (2012) LYSOZYME INTERACTION WITH NEGATIVELY CHARGED LIPID BILAYERS: PROTEIN AGGREGATION AND MEMBRANE FUSION.SOFT MATTER. VOL. 8. ISSUE 16. P. 4524-4534 | 14 | 28% | 7 |
| 2 | KONDRATYEV, KY , FEDCHENKO, PP , (2004) SOLAR RADIATION SPECTRUM AND BIOSPHERIC EVOLUTION.NUOVO CIMENTO DELLA SOCIETA ITALIANA DI FISICA C-GEOPHYSICS AND SPACE PHYSICS. VOL. 27. ISSUE 3. P. 255-272 | 6 | 75% | 0 |
| 3 | MELO, AM , RICARDO, JC , FEDOROV, A , PRIETO, M , COUTINHO, A , (2013) FLUORESCENCE DETECTION OF LIPID-INDUCED OLIGOMERIC INTERMEDIATES INVOLVED IN LYSOZYME "AMYLOID-LIKE" FIBER FORMATION DRIVEN BY ANIONIC MEMBRANES.JOURNAL OF PHYSICAL CHEMISTRY B. VOL. 117. ISSUE 10. P. 2906-2917 | 13 | 21% | 4 |
| 4 | GORBENKO, GP , (1999) FLUORESCENCE ENERGY TRANSFER STUDY OF LYSOZYME COMPLEXES WITH LIPOSOMES.BIOFIZIKA. VOL. 44. ISSUE 2. P. 257-262 | 6 | 67% | 0 |
| 5 | ZSCHORNIG, O , PAASCHE, G , THIEME, C , KORB, N , ARNOLD, K , (2005) MODULATION OF LYSOZYME CHARGE INFLUENCES INTERACTION WITH PHOSPHOLIPID VESICLES.COLLOIDS AND SURFACES B-BIOINTERFACES. VOL. 42. ISSUE 1. P. 69-78 | 8 | 38% | 31 |
| 6 | KONDRATYEV, KY , MISHEV, DN , FEDTCHENKO, PP , (1991) FORM OF THE SOLAR SPECTRUM AND BIOPROCESSES ON EARTH.DOKLADI NA BOLGARSKATA AKADEMIYA NA NAUKITE. VOL. 44. ISSUE 11. P. 39-40 | 5 | 100% | 0 |
| 7 | KONDRATYEV, KY , FEDCHENKO, PP , IVANENKO, AI , LACASA, J , FEDCHENKO, KP , (2005) ON THE BIOLOGICAL ROLE OF FRAUNHOFER LINES OF THE SUN.NUOVO CIMENTO DELLA SOCIETA ITALIANA DI FISICA C-COLLOQUIA ON PHYSICS. VOL. 28. ISSUE 6. P. 913-923 | 3 | 100% | 0 |
| 8 | MELO, AM , LOURA, LMS , FERNANDES, F , VILLALAIN, J , PRIETO, M , COUTINHO, A , (2014) ELECTROSTATICALLY DRIVEN LIPID-LYSOZYME MIXED FIBERS DISPLAY A MULTILAMELLAR STRUCTURE WITHOUT AMYLOID FEATURES.SOFT MATTER. VOL. 10. ISSUE 6. P. 840-850 | 11 | 21% | 2 |
| 9 | KHUTORSKY, VE , (1985) MODELING OF THE FORMATION OF VALINOMYCIN POTASSIUM-ION COMPLEX AT THE MEMBRANE WATER INTERFACE.BIOLOGICHESKIE MEMBRANY. VOL. 2. ISSUE 8. P. 832-843 | 6 | 100% | 1 |
| 10 | GORBENKO, GP , (1999) STRUCTURAL CHANGES IN MODEL MEMBRANES UNDER THE INFLUENCE OF RIBONUCLEASE AND LYSOZYME AS REVEALED BY FLUORESCENT PROBES.BIOFIZIKA. VOL. 44. ISSUE 6. P. 1048 -1053 | 7 | 44% | 0 |
Classes with closest relation at Level 1 |