Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
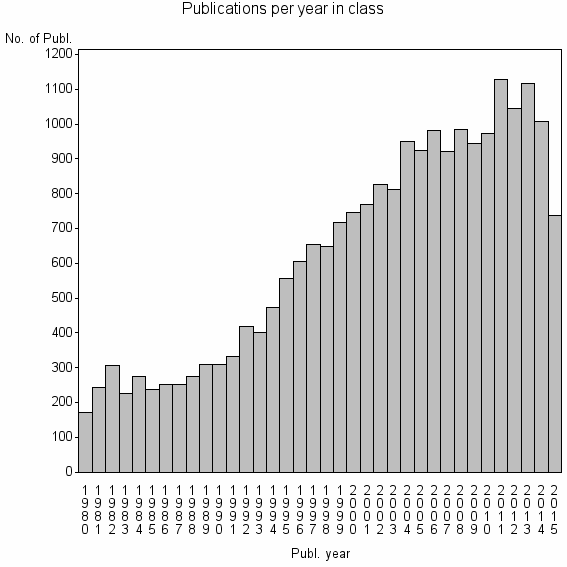
| 460 | 22518 | 44.3 | 81% |
Classes in level above (level 4) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 0 | 3633673 | BIOCHEMISTRY & MOLECULAR BIOLOGY//CELL BIOLOGY//MED |
Classes in level below (level 2) |
| ID, lev. below |
Publications | Label for level below |
|---|---|---|
| 1119 | 9219 | BACTERIORHODOPSIN//PHOTOCYCLE//PHYTOCHROME |
| 1202 | 8797 | RGS PROTEINS//G PROTEIN COUPLED RECEPTOR//RGS |
| 2168 | 4502 | CELL FREE PROTEIN SYNTHESIS//MEMBRANE PROTEIN//RIBOSOME DISPLAY |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | BACTERIORHODOPSIN | Author keyword | 722 | 67% | 3% | 659 |
| 2 | PHOTOCYCLE | Author keyword | 279 | 87% | 1% | 136 |
| 3 | PHYTOCHROME | Author keyword | 198 | 39% | 2% | 396 |
| 4 | CELL FREE PROTEIN SYNTHESIS | Author keyword | 178 | 64% | 1% | 172 |
| 5 | PHOTOTROPIN | Author keyword | 151 | 82% | 0% | 89 |
| 6 | PURPLE MEMBRANE | Author keyword | 116 | 67% | 0% | 104 |
| 7 | RHODOPSIN | Author keyword | 112 | 31% | 1% | 301 |
| 8 | RGS PROTEINS | Author keyword | 107 | 77% | 0% | 72 |
| 9 | G PROTEIN COUPLED RECEPTOR | Author keyword | 101 | 18% | 2% | 521 |
| 10 | CHLOROPLAST MOVEMENT | Author keyword | 94 | 80% | 0% | 59 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | BACTERIORHODOPSIN | 722 | 67% | 3% | 659 | Search BACTERIORHODOPSIN | Search BACTERIORHODOPSIN |
| 2 | PHOTOCYCLE | 279 | 87% | 1% | 136 | Search PHOTOCYCLE | Search PHOTOCYCLE |
| 3 | PHYTOCHROME | 198 | 39% | 2% | 396 | Search PHYTOCHROME | Search PHYTOCHROME |
| 4 | CELL FREE PROTEIN SYNTHESIS | 178 | 64% | 1% | 172 | Search CELL+FREE+PROTEIN+SYNTHESIS | Search CELL+FREE+PROTEIN+SYNTHESIS |
| 5 | PHOTOTROPIN | 151 | 82% | 0% | 89 | Search PHOTOTROPIN | Search PHOTOTROPIN |
| 6 | PURPLE MEMBRANE | 116 | 67% | 0% | 104 | Search PURPLE+MEMBRANE | Search PURPLE+MEMBRANE |
| 7 | RHODOPSIN | 112 | 31% | 1% | 301 | Search RHODOPSIN | Search RHODOPSIN |
| 8 | RGS PROTEINS | 107 | 77% | 0% | 72 | Search RGS+PROTEINS | Search RGS+PROTEINS |
| 9 | G PROTEIN COUPLED RECEPTOR | 101 | 18% | 2% | 521 | Search G+PROTEIN+COUPLED+RECEPTOR | Search G+PROTEIN+COUPLED+RECEPTOR |
| 10 | CHLOROPLAST MOVEMENT | 94 | 80% | 0% | 59 | Search CHLOROPLAST+MOVEMENT | Search CHLOROPLAST+MOVEMENT |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | PHOTOCYCLE | 1339 | 83% | 3% | 753 |
| 2 | PURPLE MEMBRANE | 706 | 62% | 3% | 733 |
| 3 | METARHODOPSIN II | 679 | 88% | 1% | 321 |
| 4 | BOVINE RHODOPSIN | 645 | 63% | 3% | 650 |
| 5 | BETA2 ADRENERGIC RECEPTOR | 618 | 37% | 6% | 1360 |
| 6 | HALOBACTERIUM HALOBIUM | 611 | 60% | 3% | 659 |
| 7 | BACTERIORHODOPSIN | 523 | 39% | 5% | 1061 |
| 8 | ASPARTIC ACID 96 | 511 | 95% | 1% | 166 |
| 9 | RHODOPSIN | 449 | 35% | 5% | 1034 |
| 10 | N INTERMEDIATE | 393 | 94% | 1% | 136 |
Journals |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | BIOPHYSICS OF STRUCTURE AND MECHANISM | 2 | 13% | 0% | 13 |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Molecular signatures of G-protein-coupled receptors | 2013 | 270 | 95 | 84% |
| The structure and function of G-protein-coupled receptors | 2009 | 623 | 68 | 82% |
| Structure-Function of the G Protein-Coupled Receptor Superfamily | 2013 | 160 | 115 | 85% |
| Molecular Mechanism of beta-Arrestin-Biased Agonism at Seven-Transmembrane Receptors | 2012 | 163 | 82 | 83% |
| Teaching old receptors new tricks: biasing seven-transmembrane receptors | 2010 | 306 | 109 | 65% |
| Microbial and Animal Rhodopsins: Structures, Functions, and Molecular Mechanisms | 2014 | 36 | 521 | 75% |
| beta-arrestins and cell signaling | 2007 | 555 | 123 | 72% |
| Diversity and modularity of G protein-coupled receptor structures | 2012 | 142 | 79 | 76% |
| Seven-transmembrane receptors | 2002 | 1168 | 113 | 57% |
| The importance of ligands for G protein-coupled receptor stability | 2015 | 4 | 56 | 84% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | ZENTRUM BIOPHYS BIOINFORMAT | 33 | 77% | 0.1% | 23 |
| 2 | MEMBRANE PROT STRUCT FUNCT UNIT | 33 | 100% | 0.1% | 13 |
| 3 | UMR 7099 | 28 | 47% | 0.2% | 45 |
| 4 | BIOL EXPT BIOPHYS | 28 | 69% | 0.1% | 24 |
| 5 | RALPH MURIEL ROBERTS VIS SCI | 26 | 87% | 0.1% | 13 |
| 6 | MOL PHARMACOL GRP | 26 | 25% | 0.4% | 91 |
| 7 | LASER BIOSCI | 24 | 91% | 0.0% | 10 |
| 8 | MOL SIGNAL TRANSDUCT SECT | 24 | 68% | 0.1% | 21 |
| 9 | MED COMPUTAC | 23 | 40% | 0.2% | 46 |
| 10 | BIOL REGULAT PHOTOBIOL | 23 | 72% | 0.1% | 18 |
Related classes at same level (level 3) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000004784 | ROLIPRAM//RHO KINASE//PHOSPHODIESTERASE |
| 2 | 0.0000003488 | PROTEIN FOLDING//PROTEINS-STRUCTURE FUNCTION AND BIOINFORMATICS//PROTEIN STRUCTURE PREDICTION |
| 3 | 0.0000003131 | RETINA//ELECTRIC FISH//ELECTRIC ORGAN DISCHARGE |
| 4 | 0.0000002296 | F 1 ATPASE//ATP SYNTHASE//V ATPASE |
| 5 | 0.0000002027 | PHAGE DISPLAY//CATALYTIC ANTIBODIES//CATALYTIC ANTIBODY |
| 6 | 0.0000001886 | JOURNAL OF MAGNETIC RESONANCE//JOURNAL OF BIOMOLECULAR NMR//JOURNAL OF MAGNETIC RESONANCE SERIES A |
| 7 | 0.0000001878 | QUORUM SENSING//PSEUDOMONAS AERUGINOSA//QUORUM QUENCHING |
| 8 | 0.0000001698 | CAFFEINE//ADENOSINE//MOL RECOGNIT SECT |
| 9 | 0.0000001622 | CLICK CHEMISTRY//SOLID PHASE SYNTHESIS//FOLDAMERS |
| 10 | 0.0000001614 | LANGMUIR//JOURNAL OF COLLOID AND INTERFACE SCIENCE//LANGMUIR BLODGETT FILMS |