Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
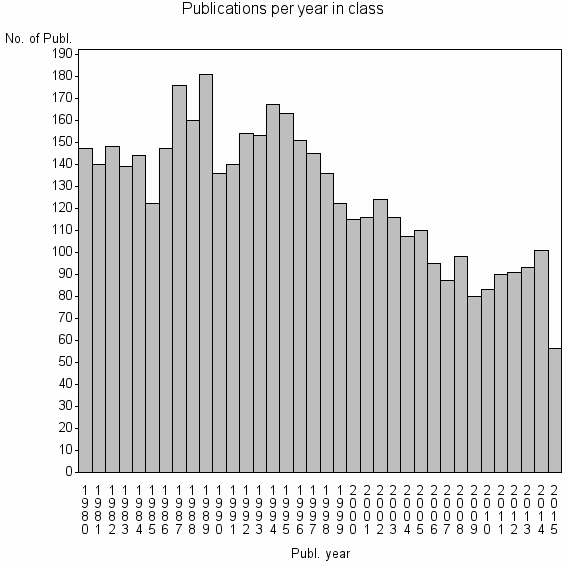
| 2150 | 4533 | 30.9 | 55% |
Classes in level above (level 3) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 295 | 37066 | CORYNEBACTERIUM GLUTAMICUM//ACTA CRYSTALLOGRAPHICA SECTION D-BIOLOGICAL CRYSTALLOGRAPHY//DIHYDRODIPICOLINATE SYNTHASE |
Classes in level below (level 1) |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | 3 ISOPROPYLMALATE DEHYDROGENASE | Author keyword | 104 | 90% | 1% | 45 |
| 2 | GLUTAMATE DEHYDROGENASE | Author keyword | 49 | 24% | 4% | 181 |
| 3 | ISOCITRATE DEHYDROGENASE | Author keyword | 47 | 33% | 3% | 116 |
| 4 | ALPHA AMINOADIPATE PATHWAY | Author keyword | 30 | 100% | 0% | 12 |
| 5 | ISOPROPYLMALATE DEHYDROGENASE | Author keyword | 27 | 78% | 0% | 18 |
| 6 | AMINO ACID DEHYDROGENASE | Author keyword | 22 | 81% | 0% | 13 |
| 7 | ISOCITRATE LYASE | Author keyword | 19 | 30% | 1% | 53 |
| 8 | THERMUS | Author keyword | 19 | 39% | 1% | 38 |
| 9 | COENZYME SPECIFICITY | Author keyword | 17 | 43% | 1% | 31 |
| 10 | HOMOISOCITRATE DEHYDROGENASE | Author keyword | 17 | 100% | 0% | 8 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | 3 ISOPROPYLMALATE DEHYDROGENASE | 104 | 90% | 1% | 45 | Search 3+ISOPROPYLMALATE+DEHYDROGENASE | Search 3+ISOPROPYLMALATE+DEHYDROGENASE |
| 2 | GLUTAMATE DEHYDROGENASE | 49 | 24% | 4% | 181 | Search GLUTAMATE+DEHYDROGENASE | Search GLUTAMATE+DEHYDROGENASE |
| 3 | ISOCITRATE DEHYDROGENASE | 47 | 33% | 3% | 116 | Search ISOCITRATE+DEHYDROGENASE | Search ISOCITRATE+DEHYDROGENASE |
| 4 | ALPHA AMINOADIPATE PATHWAY | 30 | 100% | 0% | 12 | Search ALPHA+AMINOADIPATE+PATHWAY | Search ALPHA+AMINOADIPATE+PATHWAY |
| 5 | ISOPROPYLMALATE DEHYDROGENASE | 27 | 78% | 0% | 18 | Search ISOPROPYLMALATE+DEHYDROGENASE | Search ISOPROPYLMALATE+DEHYDROGENASE |
| 6 | AMINO ACID DEHYDROGENASE | 22 | 81% | 0% | 13 | Search AMINO+ACID+DEHYDROGENASE | Search AMINO+ACID+DEHYDROGENASE |
| 7 | ISOCITRATE LYASE | 19 | 30% | 1% | 53 | Search ISOCITRATE+LYASE | Search ISOCITRATE+LYASE |
| 8 | THERMUS | 19 | 39% | 1% | 38 | Search THERMUS | Search THERMUS |
| 9 | COENZYME SPECIFICITY | 17 | 43% | 1% | 31 | Search COENZYME+SPECIFICITY | Search COENZYME+SPECIFICITY |
| 10 | HOMOISOCITRATE DEHYDROGENASE | 17 | 100% | 0% | 8 | Search HOMOISOCITRATE+DEHYDROGENASE | Search HOMOISOCITRATE+DEHYDROGENASE |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | CLOSTRIDIUM SYMBIOSUM | 113 | 91% | 1% | 48 |
| 2 | ISOPROPYLMALATE DEHYDROGENASE | 90 | 87% | 1% | 45 |
| 3 | EXTREME THERMOPHILE | 58 | 33% | 3% | 148 |
| 4 | 3 ISOPROPYLMALATE DEHYDROGENASE | 57 | 52% | 2% | 78 |
| 5 | SYMBIOSUM | 42 | 94% | 0% | 15 |
| 6 | LEUCINE DEHYDROGENASE | 33 | 71% | 1% | 27 |
| 7 | OXIDATIVE DECARBOXYLASES | 27 | 72% | 0% | 21 |
| 8 | ALPHA AMINOADIPATE PATHWAY | 26 | 61% | 1% | 27 |
| 9 | GENUS THERMUS | 25 | 63% | 1% | 25 |
| 10 | AMINO ACID DEHYDROGENASES | 23 | 100% | 0% | 10 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| The Dynamical Nature of Enzymatic Catalysis | 2015 | 2 | 34 | 56% |
| Halophilic adaptation of enzymes | 2000 | 221 | 53 | 55% |
| The alpha-aminoadipate pathway for lysine biosynthesis in fungi | 2006 | 53 | 85 | 68% |
| Malate dehydrogenases - Structure and function | 2002 | 79 | 29 | 79% |
| Molecular bases of protein halotolerance | 2014 | 5 | 76 | 41% |
| MALATE-DEHYDROGENASE - A MODEL FOR STRUCTURE, EVOLUTION, AND CATALYSIS | 1994 | 110 | 39 | 82% |
| Regulation of acetate metabolism by protein phosphorylation in enteric bacteria | 1998 | 110 | 153 | 55% |
| Untangling the glutamate dehydrogenase allosteric nightmare | 2008 | 36 | 38 | 45% |
| The structure and allosteric regulation of mammalian glutamate dehydrogenase | 2012 | 20 | 58 | 31% |
| Higher plant NADP(+)-dependent isocitrate dehydrogenases, ammonium assimilation and NADPH production | 2003 | 71 | 42 | 50% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | MOL EVOLUT BIO ERS | 4 | 29% | 0.3% | 12 |
| 2 | TG BIOTECH CO LTD | 2 | 44% | 0.1% | 4 |
| 3 | PL MOL MICROBIOL | 2 | 22% | 0.2% | 9 |
| 4 | OFF VP | 2 | 36% | 0.1% | 5 |
| 5 | SMT CHM | 2 | 67% | 0.0% | 2 |
| 6 | SOIL EROS DRY LAND FARMING LOESS P | 2 | 67% | 0.0% | 2 |
| 7 | MICROBIAL MOL PHYSIOL | 2 | 50% | 0.1% | 3 |
| 8 | BIOL STRUCT BIOPHYS MOL | 2 | 33% | 0.1% | 4 |
| 9 | CANADA GRP PROT STRUCT FUNCT | 1 | 38% | 0.1% | 3 |
| 10 | ANALYT BIOCHEM MICROBIOL | 1 | 100% | 0.0% | 2 |
Related classes at same level (level 2) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000015936 | PSEUDOMONAS SP 101//APANTELES GALLERIAE//FORMATE DEHYDROGENASE |
| 2 | 0.0000013786 | G6PD DEFICIENCY//GLUCOSE 6 PHOSPHATE DEHYDROGENASE DEFICIENCY//GLUCOSE 6 PHOSPHATE DEHYDROGENASE |
| 3 | 0.0000013408 | PHOSPHOFRUCTOKINASE//GLYCERALDEHYDE 3 PHOSPHATE DEHYDROGENASE//6 PHOSPHOFRUCTO 2 KINASE |
| 4 | 0.0000012434 | PEROXIN//PEROXISOME BIOGENESIS//PEROXISOME |
| 5 | 0.0000012143 | HYDANTOINASE//D HYDANTOINASE//DIRECTED EVOLUTION |
| 6 | 0.0000010284 | NIFA PROTEIN//GLNK//SIGMA54 |
| 7 | 0.0000010031 | ALANINE RACEMASE//TYROSINE PHENOL LYASE//MURD |
| 8 | 0.0000010013 | MACROENZYME//MACROENZYMES//MACRO CREATINE KINASE |
| 9 | 0.0000009779 | TRANSHYDROGENASE//WLDS//NAD KINASE |
| 10 | 0.0000009649 | ALKALINE PROTEASES//KERATINASE//FIBRINOLYTIC ENZYME |