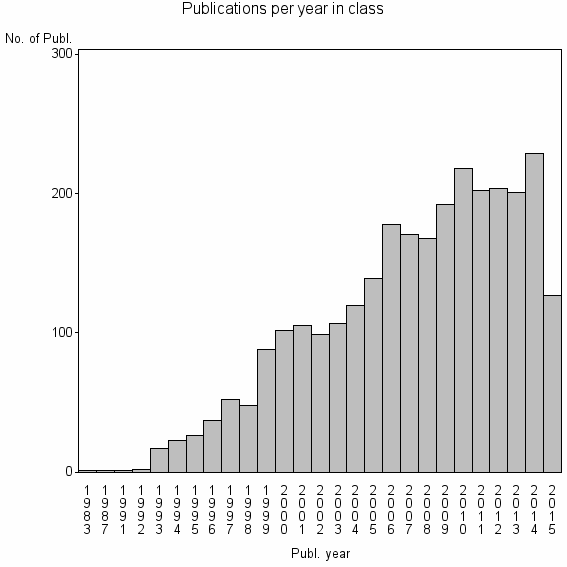
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 947 | 2858 | 51.1 | 94% |
Classes in level above (level 2) |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | MDM2 | Author keyword | 260 | 38% | 19% | 552 |
| 2 | MDMX | Author keyword | 186 | 86% | 3% | 94 |
| 3 | NUTLIN 3 | Author keyword | 69 | 64% | 2% | 68 |
| 4 | MDM4 | Author keyword | 63 | 71% | 2% | 50 |
| 5 | NUTLIN | Author keyword | 59 | 78% | 1% | 39 |
| 6 | HDM2 | Author keyword | 52 | 52% | 2% | 71 |
| 7 | HDMX | Author keyword | 44 | 88% | 1% | 21 |
| 8 | PRIMA 1 | Author keyword | 30 | 69% | 1% | 25 |
| 9 | P53 MDM2 INTERACTION | Author keyword | 23 | 79% | 1% | 15 |
| 10 | P53 | Address | 23 | 51% | 1% | 32 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | MDM2 | 260 | 38% | 19% | 552 | Search MDM2 | Search MDM2 |
| 2 | MDMX | 186 | 86% | 3% | 94 | Search MDMX | Search MDMX |
| 3 | NUTLIN 3 | 69 | 64% | 2% | 68 | Search NUTLIN+3 | Search NUTLIN+3 |
| 4 | MDM4 | 63 | 71% | 2% | 50 | Search MDM4 | Search MDM4 |
| 5 | NUTLIN | 59 | 78% | 1% | 39 | Search NUTLIN | Search NUTLIN |
| 6 | HDM2 | 52 | 52% | 2% | 71 | Search HDM2 | Search HDM2 |
| 7 | HDMX | 44 | 88% | 1% | 21 | Search HDMX | Search HDMX |
| 8 | PRIMA 1 | 30 | 69% | 1% | 25 | Search PRIMA+1 | Search PRIMA+1 |
| 9 | P53 MDM2 INTERACTION | 23 | 79% | 1% | 15 | Search P53+MDM2+INTERACTION | Search P53+MDM2+INTERACTION |
| 10 | CP 31398 | 19 | 76% | 0% | 13 | Search CP+31398 | Search CP+31398 |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | MDM2 DEFICIENT MICE | 227 | 60% | 9% | 245 |
| 2 | ONCOPROTEIN MDM2 | 214 | 64% | 7% | 209 |
| 3 | MDM2 | 124 | 23% | 16% | 468 |
| 4 | P53 MDM2 INTERACTION | 96 | 73% | 3% | 74 |
| 5 | SUPPRESSOR TRANSACTIVATION DOMAIN | 76 | 86% | 1% | 38 |
| 6 | P53 ASSOCIATED PROTEIN | 72 | 93% | 1% | 27 |
| 7 | TUMOR SUPPRESSOR P53 | 71 | 20% | 11% | 319 |
| 8 | CORE DOMAIN | 68 | 39% | 5% | 138 |
| 9 | P53 BINDING PROTEIN | 66 | 74% | 2% | 50 |
| 10 | P53 PATHWAY | 64 | 34% | 5% | 154 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| MDM2, MDMX and p53 in oncogenesis and cancer therapy | 2013 | 153 | 171 | 68% |
| Modes of p53 Regulation | 2009 | 616 | 142 | 59% |
| Blinded by the Light: The Growing Complexity of p53 | 2009 | 1004 | 220 | 35% |
| Drugging the p53 pathway: understanding the route to clinical efficacy | 2014 | 29 | 229 | 61% |
| Pharmacological reactivation of p53 as a strategy to treat cancer | 2015 | 4 | 92 | 62% |
| The p53 orchestra: Mdm2 and Mdmx set the tone | 2010 | 174 | 126 | 66% |
| MDM2 inhibitors for cancer therapy | 2007 | 289 | 80 | 80% |
| Regulating the p53 pathway: in vitro hypotheses, in vivo veritas | 2006 | 656 | 156 | 56% |
| p53 in health and disease | 2007 | 917 | 98 | 27% |
| Small-Molecule Inhibitors of the MDM2-p53 Protein-Protein Interaction to Reactivate p53 Function: A Novel Approach for Cancer Therapy | 2009 | 240 | 97 | 75% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | P53 | 23 | 51% | 1.1% | 32 |
| 2 | TRANSCRIPT NETWORKS | 10 | 52% | 0.5% | 14 |
| 3 | GRP P53 MDM2 | 6 | 80% | 0.1% | 4 |
| 4 | MCARDLE CANC 207A | 6 | 100% | 0.1% | 4 |
| 5 | STATE KAY MOL REACT DYNAM | 6 | 100% | 0.1% | 4 |
| 6 | CRUK SIGNAL TRANSDUCT GRP P53 | 5 | 55% | 0.2% | 6 |
| 7 | UNIT MOL MUTAGENESIS DNA REPAIR | 4 | 67% | 0.1% | 4 |
| 8 | CAMBRIDGE PROT ENGN | 4 | 27% | 0.5% | 14 |
| 9 | CHROMOSOME STABIL GRP | 4 | 36% | 0.3% | 8 |
| 10 | CHROMOSOME STABIL SECT | 3 | 37% | 0.2% | 7 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000287161 | MUTANT P53//GAIN OF FUNCTION//CANC THER EUT S |
| 2 | 0.0000164536 | P53 GENE//P53 MUTANTS//MUTANT P53 |
| 3 | 0.0000148239 | IASPP//ASPP2//53BP2 |
| 4 | 0.0000124227 | P53 CODON 72//P53 POLYMORPHISM//CODON 72 POLYMORPHISM |
| 5 | 0.0000108106 | VRK1//VRK2//EXPT THER EUT TRANSLAT ONCOL PROGRAM |
| 6 | 0.0000104377 | P73//P63//DELTA NP73 |
| 7 | 0.0000074091 | HIPK2//CACYBP SIP//CACYBP |
| 8 | 0.0000066244 | PPM1D//WIP1//CELL CYCLE CONTROL TUMORIGENESIS GRP |
| 9 | 0.0000063942 | PROTEOMIMETICS//ALPHA HELIX MIMETICS//HELIX MIMETICS |
| 10 | 0.0000063537 | SESTRIN 2//TP53INP1//SESTRIN |