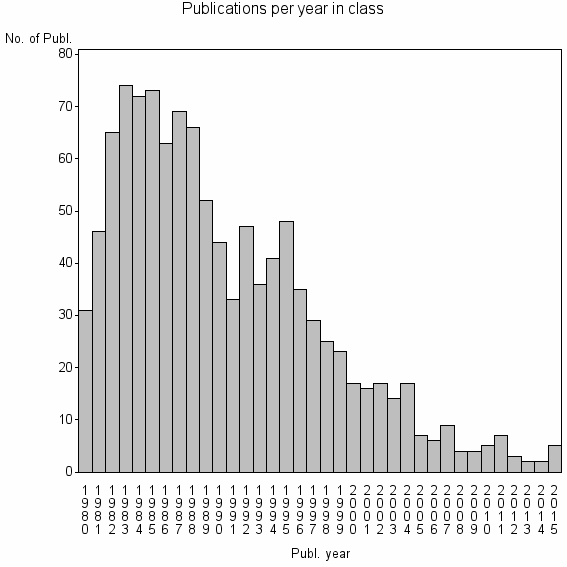
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 9222 | 1107 | 37.5 | 60% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 583 | 13522 | JOURNAL OF MAGNETIC RESONANCE//JOURNAL OF BIOMOLECULAR NMR//RESIDUAL DIPOLAR COUPLINGS |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | SHEARED G CENTER DOT A PAIR | Author keyword | 8 | 100% | 0% | 5 |
| 2 | DGGTATACC2 | Author keyword | 3 | 100% | 0% | 3 |
| 3 | SHEARED A CENTER DOT C PAIR | Author keyword | 3 | 100% | 0% | 3 |
| 4 | SHEARED GA PAIRING | Author keyword | 3 | 100% | 0% | 3 |
| 5 | HUMAN CENTROMERE | Author keyword | 2 | 67% | 0% | 2 |
| 6 | PROTON CHEMICAL SHIFT | Author keyword | 1 | 31% | 0% | 4 |
| 7 | A FORM CONFORMATION | Author keyword | 1 | 100% | 0% | 2 |
| 8 | BASE INTERCALATED DUPLEX | Author keyword | 1 | 100% | 0% | 2 |
| 9 | DNA BULGES | Author keyword | 1 | 50% | 0% | 2 |
| 10 | G CENTER DOT A MISMATCH | Author keyword | 1 | 50% | 0% | 2 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | SHEARED G CENTER DOT A PAIR | 8 | 100% | 0% | 5 | Search SHEARED+G+CENTER+DOT+A+PAIR | Search SHEARED+G+CENTER+DOT+A+PAIR |
| 2 | DGGTATACC2 | 3 | 100% | 0% | 3 | Search DGGTATACC2 | Search DGGTATACC2 |
| 3 | SHEARED A CENTER DOT C PAIR | 3 | 100% | 0% | 3 | Search SHEARED+A+CENTER+DOT+C+PAIR | Search SHEARED+A+CENTER+DOT+C+PAIR |
| 4 | SHEARED GA PAIRING | 3 | 100% | 0% | 3 | Search SHEARED+GA+PAIRING | Search SHEARED+GA+PAIRING |
| 5 | HUMAN CENTROMERE | 2 | 67% | 0% | 2 | Search HUMAN+CENTROMERE | Search HUMAN+CENTROMERE |
| 6 | PROTON CHEMICAL SHIFT | 1 | 31% | 0% | 4 | Search PROTON+CHEMICAL+SHIFT | Search PROTON+CHEMICAL+SHIFT |
| 7 | A FORM CONFORMATION | 1 | 100% | 0% | 2 | Search A+FORM+CONFORMATION | Search A+FORM+CONFORMATION |
| 8 | BASE INTERCALATED DUPLEX | 1 | 100% | 0% | 2 | Search BASE+INTERCALATED+DUPLEX | Search BASE+INTERCALATED+DUPLEX |
| 9 | DNA BULGES | 1 | 50% | 0% | 2 | Search DNA+BULGES | Search DNA+BULGES |
| 10 | G CENTER DOT A MISMATCH | 1 | 50% | 0% | 2 | Search G+CENTER+DOT+A+MISMATCH | Search G+CENTER+DOT+A+MISMATCH |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | GA MISMATCH | 24 | 60% | 2% | 26 |
| 2 | CONTAINING DEOXYTRIDECANUCLEOTIDE DUPLEXES | 20 | 52% | 3% | 28 |
| 3 | ABASIC FRAMESHIFT | 15 | 88% | 1% | 7 |
| 4 | ADENOSINE STACKS | 12 | 75% | 1% | 9 |
| 5 | EXTRA ADENOSINE STACKS | 12 | 59% | 1% | 13 |
| 6 | DNA OCTAMER DM5C G M5C G T G M5C G | 11 | 100% | 1% | 6 |
| 7 | FLANKING SEQUENCE | 10 | 42% | 2% | 19 |
| 8 | RESIDUE A5 | 9 | 83% | 0% | 5 |
| 9 | OCTAMER DM5C G M5C G T G M5C G | 8 | 100% | 0% | 5 |
| 10 | CENTER DOT A | 8 | 30% | 2% | 22 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| KINKING OF DNA AND RNA BY BASE BULGES | 1995 | 101 | 29 | 72% |
| Use of chemical shifts for structural studies of nucleic acids | 2010 | 19 | 137 | 52% |
| Unusual DNA duplex and hairpin motifs | 2003 | 57 | 117 | 33% |
| NUCLEIC-ACIDS AND NUCLEAR MAGNETIC-RESONANCE | 1988 | 286 | 353 | 56% |
| CONFORMATION AND DYNAMICS OF DNA AND PROTEIN-DNA COMPLEXES BY P-31 NMR | 1994 | 169 | 165 | 36% |
| P-31 NMR OF DNA | 1992 | 78 | 60 | 58% |
| DNA AND RNA - NMR-STUDIES OF CONFORMATIONS AND DYNAMICS IN SOLUTION | 1987 | 148 | 143 | 59% |
| A zipper-like duplex in DNA: the crystal structure of d(GCGAAAGCT) at 2.1 angstrom resolution | 1998 | 45 | 86 | 28% |
| Conformations of an adenine bulge in a DNA octamer and its influence on DNA structure from molecular dynamics simulations | 2001 | 23 | 95 | 27% |
| Design, synthesis, and analysis of disulfide cross-linked DNA duplexes | 1996 | 33 | 88 | 30% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | AG THEORET BIOPHYS | 1 | 50% | 0.1% | 1 |
| 2 | FRE2600 | 1 | 50% | 0.1% | 1 |
| 3 | GRP B1 | 1 | 50% | 0.1% | 1 |
| 4 | HLTH SCI DEV PROGRAM | 1 | 50% | 0.1% | 1 |
| 5 | MCCLINTOCK OURCE | 1 | 50% | 0.1% | 1 |
| 6 | UMR 8532PR2 | 1 | 50% | 0.1% | 1 |
| 7 | CRC NUCL ACID STRUCT GRP | 0 | 33% | 0.1% | 1 |
| 8 | CORE DNA SYNTH IL | 0 | 25% | 0.1% | 1 |
| 9 | CANC NANOBIOL | 0 | 20% | 0.1% | 1 |
| 10 | CNRS ESA 7033 | 0 | 20% | 0.1% | 1 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000240200 | RIBOSE AND NUCLEOBASE//ALTERNATE SITE SPECIFIC LABELING//DL323 |
| 2 | 0.0000221157 | 3J COUPLING CONSTANTS//DISTANCE GEOMETRY//TIME AVERAGED RESTRAINTS |
| 3 | 0.0000178723 | DNA CURVATURE//A TRACT//DNA FLEXIBILITY |
| 4 | 0.0000166401 | BASE PAIR KINETICS//GNRA TETRALOOPS//RAAA HAIRPIN MOTIF |
| 5 | 0.0000157158 | Z DNA//B Z TRANSITION//Z DNA BINDING PROTEIN |
| 6 | 0.0000127691 | L DNA//HETEROCHIRAL DNA//UMR CNRS UMII 5625 |
| 7 | 0.0000121958 | 5S RRNA//5S RIBOSOMAL RNA//ARBEITSGRP MAKROMOL STRUKTURANAL |
| 8 | 0.0000120345 | HETERO ASSOCIATION//HETEROASSOCIATION//ACTINOMYCINS |
| 9 | 0.0000102832 | PSEUDOROTATIONAL EQUILIBRIUM//3JHH COUPLING CONSTANTS//ABINITIO MOLECULAR ORBITAL CALCULATION |
| 10 | 0.0000095128 | PHOSPHOROTHIOLATE//P 32ORTHOPHOSPHORIC ACID//SYNTHESIS OF P 32 LABELED NUCLEOTIDES |