Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
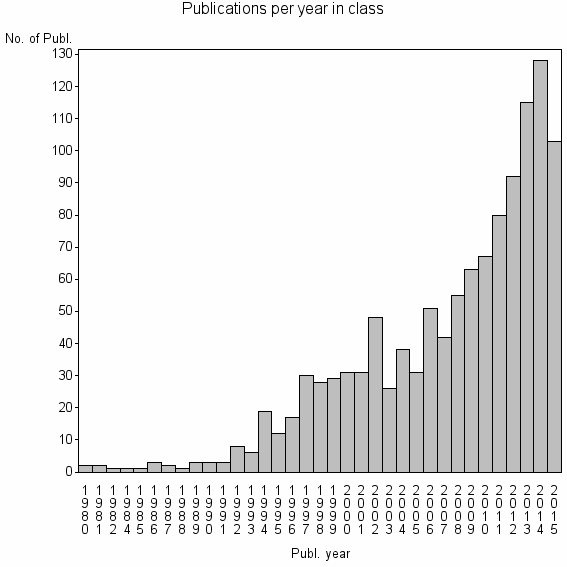
| 8570 | 1172 | 53.2 | 89% |
Classes in level above (level 2) |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | SNRK2 | Author keyword | 44 | 85% | 2% | 23 |
| 2 | ABI3 | Author keyword | 33 | 74% | 2% | 25 |
| 3 | ABA RECEPTOR | Author keyword | 32 | 88% | 1% | 15 |
| 4 | 9 CIS EPOXYCAROTENOID DIOXYGENASE NCED | Author keyword | 21 | 90% | 1% | 9 |
| 5 | ABA SIGNAL TRANSDUCTION | Author keyword | 21 | 90% | 1% | 9 |
| 6 | 9 CIS EPOXYCAROTENOID DIOXYGENASE | Author keyword | 17 | 63% | 1% | 17 |
| 7 | ABI5 | Author keyword | 15 | 54% | 2% | 19 |
| 8 | ABA SIGNALING | Author keyword | 14 | 50% | 2% | 20 |
| 9 | ABI1 | Author keyword | 12 | 56% | 1% | 15 |
| 10 | PYL | Author keyword | 12 | 86% | 1% | 6 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | SNRK2 | 44 | 85% | 2% | 23 | Search SNRK2 | Search SNRK2 |
| 2 | ABI3 | 33 | 74% | 2% | 25 | Search ABI3 | Search ABI3 |
| 3 | ABA RECEPTOR | 32 | 88% | 1% | 15 | Search ABA+RECEPTOR | Search ABA+RECEPTOR |
| 4 | 9 CIS EPOXYCAROTENOID DIOXYGENASE NCED | 21 | 90% | 1% | 9 | Search 9+CIS+EPOXYCAROTENOID+DIOXYGENASE+NCED | Search 9+CIS+EPOXYCAROTENOID+DIOXYGENASE+NCED |
| 5 | ABA SIGNAL TRANSDUCTION | 21 | 90% | 1% | 9 | Search ABA+SIGNAL+TRANSDUCTION | Search ABA+SIGNAL+TRANSDUCTION |
| 6 | 9 CIS EPOXYCAROTENOID DIOXYGENASE | 17 | 63% | 1% | 17 | Search 9+CIS+EPOXYCAROTENOID+DIOXYGENASE | Search 9+CIS+EPOXYCAROTENOID+DIOXYGENASE |
| 7 | ABI5 | 15 | 54% | 2% | 19 | Search ABI5 | Search ABI5 |
| 8 | ABA SIGNALING | 14 | 50% | 2% | 20 | Search ABA+SIGNALING | Search ABA+SIGNALING |
| 9 | ABI1 | 12 | 56% | 1% | 15 | Search ABI1 | Search ABI1 |
| 10 | PYL | 12 | 86% | 1% | 6 | Search PYL | Search PYL |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | PYL PROTEINS | 65 | 95% | 2% | 21 |
| 2 | ABSCISIC ACID BIOSYNTHESIS | 46 | 28% | 12% | 139 |
| 3 | PROTEIN PHOSPHATASE 2C | 37 | 40% | 6% | 72 |
| 4 | ABI1 | 37 | 63% | 3% | 37 |
| 5 | 9 CIS EPOXYCAROTENOID DIOXYGENASE | 36 | 43% | 5% | 63 |
| 6 | LATE EMBRYO DEVELOPMENT | 30 | 79% | 2% | 19 |
| 7 | LATE EMBRYOGENESIS | 26 | 48% | 3% | 40 |
| 8 | CONSERVED B3 DOMAIN | 25 | 70% | 2% | 21 |
| 9 | 9 CIS EPOXYCAROTENOID DIOXYGENASE GENE | 25 | 50% | 3% | 36 |
| 10 | LEAFY COTYLEDON1 | 25 | 43% | 4% | 44 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Abscisic Acid: Emergence of a Core Signaling Network | 2010 | 494 | 176 | 48% |
| ABA signaling in stress-response and seed development | 2013 | 47 | 99 | 73% |
| Pivotal role of the AREB/ABF-SnRK2 pathway in ABRE-mediated transcription in response to osmotic stress in plants | 2013 | 31 | 100 | 70% |
| ABA perception and signalling | 2010 | 211 | 84 | 60% |
| Structural basis and functions of abscisic acid receptors PYLs | 2015 | 1 | 124 | 75% |
| Molecular Basis of the Core Regulatory Network in ABA Responses: Sensing, Signaling and Transport | 2010 | 154 | 109 | 72% |
| Structure and function of abscisic acid receptors | 2013 | 21 | 79 | 72% |
| The Roles of ROS and ABA in Systemic Acquired Acclimation | 2015 | 3 | 45 | 27% |
| Abscisic acid biosynthesis and catabolism | 2005 | 619 | 125 | 33% |
| ABA-dependent and ABA-independent signaling in response to osmotic stress in plants | 2014 | 11 | 45 | 49% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | MOE SYST BIOL BIOINFORMAT | 9 | 83% | 0.4% | 5 |
| 2 | REPROD GROWTH REGULAT | 6 | 80% | 0.3% | 4 |
| 3 | LEHRSTUHL BOT | 5 | 22% | 1.9% | 22 |
| 4 | DORMANCY AD TAT UNIT | 4 | 67% | 0.3% | 4 |
| 5 | GENE DISCOVERY GRP | 4 | 19% | 1.5% | 18 |
| 6 | CELL DEV BIOL SECT | 3 | 17% | 1.6% | 19 |
| 7 | UPR 2355 | 2 | 13% | 1.4% | 16 |
| 8 | PLANT SCI PARIS SACLAY | 2 | 67% | 0.2% | 2 |
| 9 | BIOL OURCES POSTHARVEST | 2 | 17% | 0.8% | 9 |
| 10 | CULTIVAT TOBACCO IND | 1 | 50% | 0.2% | 2 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000258748 | U BOX//CELLULAR BIOL MOL GENET//PLANT BIOL GRP PROGRAM |
| 2 | 0.0000226801 | DREB//AP2 ERF//GCC BOX |
| 3 | 0.0000202484 | SNRK1//HEXOSE PHOSPHORYLATION//FRUCTOKINASE |
| 4 | 0.0000171336 | PHASEIC ACID//ABSCISIC ACID//PARTIAL ROOT DRYING |
| 5 | 0.0000158609 | ENDO BETA MANNANASE//ENDOSPERM WEAKENING//SEED DORMANCY |
| 6 | 0.0000131376 | GUARD CELL//GUARD CELLS//SV CHANNEL |
| 7 | 0.0000130999 | AGB1//HETEROTRIMERIC G PROTEIN//HETEROTRIMERIC G PROTEIN BETA SUBUNIT |
| 8 | 0.0000129076 | WRKY TRANSCRIPTION FACTOR//WRKY//WRKY PROTEIN |
| 9 | 0.0000128881 | DEHYDRIN//LEA PROTEIN//DEHYDRINS |
| 10 | 0.0000127556 | MAPKK//MAPKKK//MAPK CASCADE |