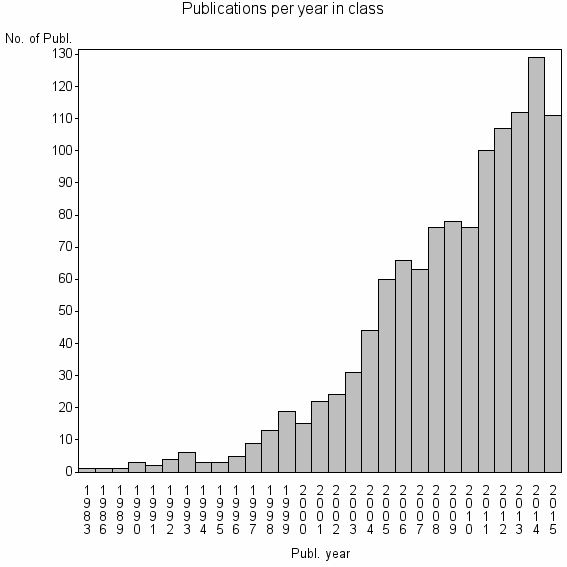
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 8439 | 1184 | 41.6 | 77% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 42 | 31225 | PROTEINS-STRUCTURE FUNCTION AND BIOINFORMATICS//PROTEIN STRUCTURE PREDICTION//PROTEIN FOLDING |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | MARKOV STATE MODELS | Author keyword | 19 | 80% | 1% | 12 |
| 2 | TRANSITION PATH SAMPLING | Author keyword | 9 | 55% | 1% | 12 |
| 3 | TRANSITION PATH THEORY | Author keyword | 9 | 83% | 0% | 5 |
| 4 | ENHANCED SAMPLING | Author keyword | 8 | 36% | 1% | 17 |
| 5 | IST SCI COMPUTAZ | Address | 7 | 43% | 1% | 13 |
| 6 | ACTION DERIVED MOLECULAR DYNAMICS | Author keyword | 6 | 80% | 0% | 4 |
| 7 | MILESTONING | Author keyword | 6 | 80% | 0% | 4 |
| 8 | METADYNAMICS | Author keyword | 6 | 26% | 2% | 19 |
| 9 | HYPER MOLECULAR DYNAMICS | Author keyword | 6 | 100% | 0% | 4 |
| 10 | MARKOV STATE MODEL | Author keyword | 5 | 47% | 1% | 8 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | MARKOV STATE MODELS | 19 | 80% | 1% | 12 | Search MARKOV+STATE+MODELS | Search MARKOV+STATE+MODELS |
| 2 | TRANSITION PATH SAMPLING | 9 | 55% | 1% | 12 | Search TRANSITION+PATH+SAMPLING | Search TRANSITION+PATH+SAMPLING |
| 3 | TRANSITION PATH THEORY | 9 | 83% | 0% | 5 | Search TRANSITION+PATH+THEORY | Search TRANSITION+PATH+THEORY |
| 4 | ENHANCED SAMPLING | 8 | 36% | 1% | 17 | Search ENHANCED+SAMPLING | Search ENHANCED+SAMPLING |
| 5 | ACTION DERIVED MOLECULAR DYNAMICS | 6 | 80% | 0% | 4 | Search ACTION+DERIVED+MOLECULAR+DYNAMICS | Search ACTION+DERIVED+MOLECULAR+DYNAMICS |
| 6 | MILESTONING | 6 | 80% | 0% | 4 | Search MILESTONING | Search MILESTONING |
| 7 | METADYNAMICS | 6 | 26% | 2% | 19 | Search METADYNAMICS | Search METADYNAMICS |
| 8 | HYPER MOLECULAR DYNAMICS | 6 | 100% | 0% | 4 | Search HYPER+MOLECULAR+DYNAMICS | Search HYPER+MOLECULAR+DYNAMICS |
| 9 | MARKOV STATE MODEL | 5 | 47% | 1% | 8 | Search MARKOV+STATE+MODEL | Search MARKOV+STATE+MODEL |
| 10 | CONFORMATION DYNAMICS | 3 | 43% | 1% | 6 | Search CONFORMATION+DYNAMICS | Search CONFORMATION+DYNAMICS |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | STRING METHOD | 45 | 55% | 5% | 57 |
| 2 | INFREQUENT EVENTS | 42 | 34% | 8% | 99 |
| 3 | METADYNAMICS | 28 | 44% | 4% | 49 |
| 4 | HYPERDYNAMICS | 27 | 78% | 2% | 18 |
| 5 | RARE EVENTS | 18 | 23% | 6% | 71 |
| 6 | PROTEIN FOLDING KINETICS | 17 | 33% | 4% | 43 |
| 7 | STATE MODELS | 17 | 52% | 2% | 23 |
| 8 | ISOCOMMITTOR SURFACES | 15 | 71% | 1% | 12 |
| 9 | ACCELERATED MOLECULAR DYNAMICS | 10 | 30% | 2% | 29 |
| 10 | ALANINE DIPEPTIDE | 9 | 17% | 4% | 48 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Practical and conceptual path sampling issues | 2015 | 2 | 52 | 92% |
| A critical appraisal of Markov state models | 2015 | 2 | 23 | 91% |
| Discovering Mountain Passes via Torchlight: Methods for the Definition of Reaction Coordinates and Pathways in Complex Macromolecular Reactions | 2013 | 21 | 107 | 58% |
| Metadynamics: a method to simulate rare events and reconstruct the free energy in biophysics, chemistry and material science | 2008 | 291 | 133 | 38% |
| Accelerated dynamics: Mathematical foundations and algorithmic improvements | 2015 | 2 | 6 | 67% |
| Metadynamics | 2011 | 111 | 119 | 34% |
| Metadynamics as a tool for exploring free energy landscapes of chemical reactions | 2006 | 155 | 40 | 60% |
| Forward flux sampling for rare event simulations | 2009 | 95 | 84 | 54% |
| Towards fast, rigorous and efficient conformational sampling of biomolecules: Advances in accelerated molecular dynamics | 2015 | 2 | 54 | 54% |
| Everything you wanted to know about Markov State Models but were afraid to ask | 2010 | 65 | 44 | 61% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | IST SCI COMPUTAZ | 7 | 43% | 1.1% | 13 |
| 2 | SIMBIOS NIH BIOMED COMPUTAT | 4 | 67% | 0.3% | 4 |
| 3 | MICMAC PROJECT TEAM | 2 | 33% | 0.5% | 6 |
| 4 | MATHEON | 2 | 44% | 0.3% | 4 |
| 5 | IST SCI COMPUTAZIONALI | 2 | 43% | 0.3% | 3 |
| 6 | COMPUTAT BIOPHYS GRIB IMIM | 1 | 38% | 0.3% | 3 |
| 7 | INTERDISCIPLINARY ADV MAT SIMULAT ICAMS | 1 | 50% | 0.2% | 2 |
| 8 | SYST BIOL HUMAN HLTH | 1 | 16% | 0.6% | 7 |
| 9 | HIGH PERFORMANCE SIMULAT CHIPS | 1 | 27% | 0.3% | 4 |
| 10 | HIGH PERFORMANCE CLUSTER COMP | 1 | 33% | 0.2% | 2 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000214849 | VALLEY RIDGE INFLECTION POINT//NEWTON TRAJECTORY//GRADIENT EXTREMAL |
| 2 | 0.0000193027 | BAKER CHEM CHEM BIOL//REPLICA EXCHANGE METHOD//GENERALIZED ENSEMBLE ALGORITHM |
| 3 | 0.0000142003 | ENVIRONM IND CHEM//ONE STEP PERTURBATION//SAMPL4 |
| 4 | 0.0000121293 | PROTEIN FOLDING//PHI VALUE//CONTACT ORDER |
| 5 | 0.0000108954 | ELASTIC NETWORK MODEL//ELASTIC NETWORK MODELS//NORMAL MODE ANALYSIS |
| 6 | 0.0000084594 | BETA HAIRPIN//SIDE CHAIN INTERACTIONS//TRP CAGE |
| 7 | 0.0000083056 | CRITICAL DROPLET//CRITICAL DROPLETS//METASTABLE TRANSITION TIME |
| 8 | 0.0000076350 | CARBONIC ACID//AUTOCATALYTIC DECOMPOSITION REACTION//PROTON TRANSFER BARRIERS |
| 9 | 0.0000071494 | BIOPHYS MODELING SIMULAT//ALL ATOM RECONSTRUCTION//BIOL MODELING SIMULAT |
| 10 | 0.0000060912 | ATMOSPHERE OCEAN SCI//MULTISCALE DIFFUSIONS//MORI ZWANZIG |