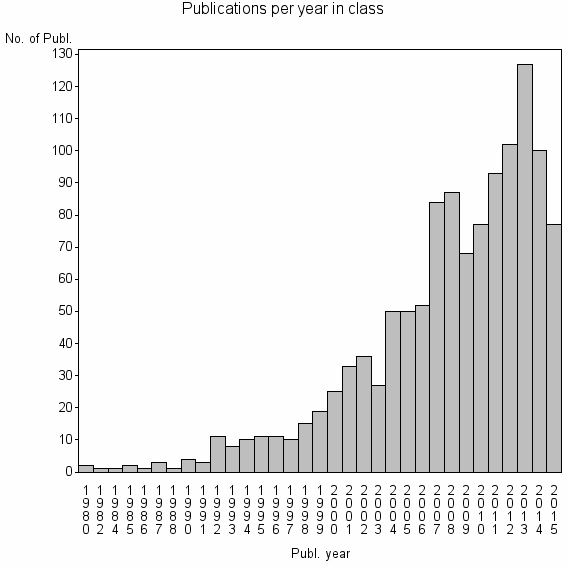
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 8272 | 1201 | 50.8 | 93% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 195 | 20594 | PRE MRNA SPLICING//RNA LOCALIZATION//SPLICEOSOME |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | DECAPPING | Author keyword | 42 | 61% | 4% | 44 |
| 2 | P BODIES | Author keyword | 34 | 50% | 4% | 50 |
| 3 | CCR4 NOT | Author keyword | 28 | 81% | 1% | 17 |
| 4 | STRESS GRANULES | Author keyword | 26 | 37% | 5% | 57 |
| 5 | MRNA DECAY | Author keyword | 21 | 26% | 6% | 71 |
| 6 | DEADENYLATION | Author keyword | 21 | 37% | 4% | 44 |
| 7 | DEADENYLASE | Author keyword | 19 | 68% | 1% | 17 |
| 8 | CCR4 NOT COMPLEX | Author keyword | 19 | 71% | 1% | 15 |
| 9 | DECAPPING ENZYME | Author keyword | 18 | 89% | 1% | 8 |
| 10 | GW BODIES | Author keyword | 18 | 89% | 1% | 8 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | DECAPPING | 42 | 61% | 4% | 44 | Search DECAPPING | Search DECAPPING |
| 2 | P BODIES | 34 | 50% | 4% | 50 | Search P+BODIES | Search P+BODIES |
| 3 | CCR4 NOT | 28 | 81% | 1% | 17 | Search CCR4+NOT | Search CCR4+NOT |
| 4 | STRESS GRANULES | 26 | 37% | 5% | 57 | Search STRESS+GRANULES | Search STRESS+GRANULES |
| 5 | MRNA DECAY | 21 | 26% | 6% | 71 | Search MRNA+DECAY | Search MRNA+DECAY |
| 6 | DEADENYLATION | 21 | 37% | 4% | 44 | Search DEADENYLATION | Search DEADENYLATION |
| 7 | DEADENYLASE | 19 | 68% | 1% | 17 | Search DEADENYLASE | Search DEADENYLASE |
| 8 | CCR4 NOT COMPLEX | 19 | 71% | 1% | 15 | Search CCR4+NOT+COMPLEX | Search CCR4+NOT+COMPLEX |
| 9 | DECAPPING ENZYME | 18 | 89% | 1% | 8 | Search DECAPPING+ENZYME | Search DECAPPING+ENZYME |
| 10 | GW BODIES | 18 | 89% | 1% | 8 | Search GW+BODIES | Search GW+BODIES |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | CYTOPLASMIC PROCESSING BODIES | 108 | 59% | 10% | 121 |
| 2 | PROCESSING BODIES | 98 | 44% | 14% | 167 |
| 3 | CAF1 PROTEINS | 83 | 82% | 4% | 49 |
| 4 | P BODIES | 72 | 37% | 13% | 155 |
| 5 | DECAPPING ENZYME | 42 | 47% | 5% | 66 |
| 6 | P BODY FORMATION | 42 | 60% | 4% | 46 |
| 7 | DEADENYLATION | 37 | 29% | 9% | 108 |
| 8 | ENDORIBONUCLEASE G3BP | 36 | 66% | 3% | 33 |
| 9 | PROCESSING BODY | 35 | 86% | 1% | 18 |
| 10 | CYTOPLASMIC FOCI | 34 | 61% | 3% | 36 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| P bodies and the control of mRNA translation and degradation | 2007 | 587 | 92 | 51% |
| Eukaryotic Stress Granules: The Ins and Outs of Translation | 2009 | 334 | 95 | 53% |
| Stress granules: The Tao of RNA triage | 2008 | 369 | 86 | 43% |
| RNA granules | 2006 | 526 | 58 | 53% |
| RNA granules: post-transcriptional and epigenetic modulators of gene expression | 2009 | 301 | 72 | 51% |
| The Control of mRNA Decapping and P-Body Formation | 2008 | 181 | 121 | 70% |
| The Ccr4-Not complex | 2012 | 78 | 176 | 52% |
| Mammalian stress granules and processing bodies | 2007 | 244 | 34 | 76% |
| RNA decay machines: Deadenylation by the Ccr4-Not and Pan2-Pan3 complexes | 2013 | 36 | 128 | 58% |
| P bodies: at the crossroads of post-transcriptional pathways | 2007 | 494 | 119 | 39% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | CNRS FRE 3402 | 4 | 67% | 0.3% | 4 |
| 2 | CELL SIGNAL UNIT | 3 | 35% | 0.5% | 6 |
| 3 | HELMHOLTZ JR GRP POSTTRANSCRIPT CONTROL GENE | 3 | 35% | 0.5% | 6 |
| 4 | BIOL CHEM MOL PHARMACOLHEMATOL | 2 | 67% | 0.2% | 2 |
| 5 | HIV RNA TRAFFICKING 1 | 2 | 29% | 0.4% | 5 |
| 6 | CNRS UMR 8122 | 1 | 50% | 0.2% | 2 |
| 7 | FRE 3402 | 1 | 50% | 0.2% | 2 |
| 8 | GLUCOSE SIGNALING GRP | 1 | 100% | 0.2% | 2 |
| 9 | TRANSLAT FOLDING TEAM | 1 | 50% | 0.2% | 2 |
| 10 | RECH HOP ST FRANCOIS DASSISE | 1 | 40% | 0.2% | 2 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000194580 | UPF1//NMD//NONSENSE MEDIATED MRNA DECAY |
| 2 | 0.0000176130 | CYTOPLASMIC POLYADENYLATION//PUMILIO//CPEB |
| 3 | 0.0000171110 | HUR//TRISTETRAPROLIN//AU RICH ELEMENT |
| 4 | 0.0000164762 | MRNP BIOGENESIS METAB//DIS3//RRP6 |
| 5 | 0.0000157472 | TOB//BTG1//BTG2 |
| 6 | 0.0000135148 | POLYA BINDING PROTEIN//POLY A BINDING PROTEIN//PABP |
| 7 | 0.0000105163 | MCPIP1//ZC3H12A//MCPIP |
| 8 | 0.0000093082 | RNA HELICASE//DEAD BOX//DEAD BOX PROTEIN |
| 9 | 0.0000082986 | EIF4E//EIF 4E//CAPPING EFFICIENCY |
| 10 | 0.0000064357 | AXONAL PROTEIN SYNTHESIS//LOCAL PROTEIN SYNTHESIS//DENDRITIC MRNA TRANSPORT |