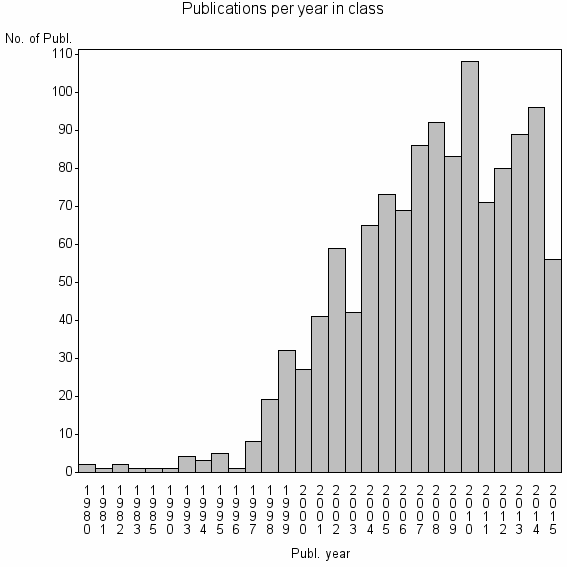
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 8125 | 1217 | 25.5 | 67% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 2779 | 2619 | GMO DETECTION//EVENT SPECIFIC//LOOP MEDIATED ISOTHERMAL AMPLIFICATION |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | GMO DETECTION | Author keyword | 247 | 98% | 5% | 60 |
| 2 | EVENT SPECIFIC | Author keyword | 114 | 95% | 3% | 38 |
| 3 | GMO | Author keyword | 61 | 31% | 13% | 161 |
| 4 | GMO DETECT | Address | 41 | 87% | 2% | 20 |
| 5 | QUALITATIVE AND QUANTITATIVE PCR | Author keyword | 34 | 93% | 1% | 13 |
| 6 | GENETICALLY MODIFIED ORGANISM | Author keyword | 34 | 38% | 6% | 71 |
| 7 | ENDOGENOUS REFERENCE GENE | Author keyword | 28 | 81% | 1% | 17 |
| 8 | GENETICALLY MODIFIED ORGANISMS | Author keyword | 28 | 27% | 7% | 90 |
| 9 | GENETICALLY MODIFIED ORGANISM GMO | Author keyword | 27 | 54% | 3% | 35 |
| 10 | EVENT SPECIFIC DETECTION | Author keyword | 24 | 91% | 1% | 10 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | GMO DETECTION | 247 | 98% | 5% | 60 | Search GMO+DETECTION | Search GMO+DETECTION |
| 2 | EVENT SPECIFIC | 114 | 95% | 3% | 38 | Search EVENT+SPECIFIC | Search EVENT+SPECIFIC |
| 3 | GMO | 61 | 31% | 13% | 161 | Search GMO | Search GMO |
| 4 | QUALITATIVE AND QUANTITATIVE PCR | 34 | 93% | 1% | 13 | Search QUALITATIVE+AND+QUANTITATIVE+PCR | Search QUALITATIVE+AND+QUANTITATIVE+PCR |
| 5 | GENETICALLY MODIFIED ORGANISM | 34 | 38% | 6% | 71 | Search GENETICALLY+MODIFIED+ORGANISM | Search GENETICALLY+MODIFIED+ORGANISM |
| 6 | ENDOGENOUS REFERENCE GENE | 28 | 81% | 1% | 17 | Search ENDOGENOUS+REFERENCE+GENE | Search ENDOGENOUS+REFERENCE+GENE |
| 7 | GENETICALLY MODIFIED ORGANISMS | 28 | 27% | 7% | 90 | Search GENETICALLY+MODIFIED+ORGANISMS | Search GENETICALLY+MODIFIED+ORGANISMS |
| 8 | GENETICALLY MODIFIED ORGANISM GMO | 27 | 54% | 3% | 35 | Search GENETICALLY+MODIFIED+ORGANISM+GMO | Search GENETICALLY+MODIFIED+ORGANISM+GMO |
| 9 | EVENT SPECIFIC DETECTION | 24 | 91% | 1% | 10 | Search EVENT+SPECIFIC+DETECTION | Search EVENT+SPECIFIC+DETECTION |
| 10 | EVENT SPECIFIC PCR | 24 | 91% | 1% | 10 | Search EVENT+SPECIFIC+PCR | Search EVENT+SPECIFIC+PCR |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | MODIFIED ORGANISMS | 336 | 82% | 16% | 193 |
| 2 | GENETICALLY MODIFIED ORGANISMS | 229 | 67% | 17% | 209 |
| 3 | MODIFIED MAIZE | 198 | 81% | 10% | 118 |
| 4 | REFERENCE MOLECULES | 140 | 86% | 6% | 70 |
| 5 | EVENT SPECIFIC DETECTION | 89 | 92% | 3% | 35 |
| 6 | GMO | 83 | 84% | 4% | 46 |
| 7 | ENDOGENOUS REFERENCE GENE | 52 | 82% | 3% | 31 |
| 8 | GMOS | 52 | 71% | 3% | 42 |
| 9 | QUANTITATIVE PCR DETECTION | 41 | 87% | 2% | 20 |
| 10 | DETECTING RECOMBINANT DNAS | 41 | 100% | 1% | 15 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Influence of DNA extraction methods, PCR inhibitors and quantification methods on real-time PCR assay of biotechnology-derived traits | 2010 | 73 | 43 | 72% |
| Guidelines for validation of qualitative real-time PCR methods | 2014 | 6 | 23 | 74% |
| The Development and Standardization of Testing Methods for Genetically Modified Organisms and their Derived Products | 2011 | 24 | 90 | 82% |
| Detecting un-authorized genetically modified organisms (GMOs) and derived materials | 2012 | 16 | 60 | 85% |
| Effect of food processing on plant DNA degradation and PCR-based GMO analysis: a review | 2010 | 44 | 116 | 77% |
| New approaches in GMO detection | 2010 | 28 | 30 | 97% |
| Methods for detection of GMOs in food and feed | 2008 | 68 | 78 | 59% |
| Genetically modified crops: Detection strategies and biosafety issues | 2013 | 14 | 35 | 57% |
| Advances in molecular techniques for the detection and quantification of genetically modified organisms | 2008 | 51 | 45 | 69% |
| Detection of genetically modified organisms in foods | 2002 | 182 | 30 | 67% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | GMO DETECT | 41 | 87% | 1.6% | 20 |
| 2 | BIOTECHNOL GMOS UNIT | 21 | 85% | 0.9% | 11 |
| 3 | MOL CHARACTERIZAT GENETICALLY MODIFIED O | 14 | 64% | 1.2% | 14 |
| 4 | METHODOL DETECT OGM | 14 | 100% | 0.6% | 7 |
| 5 | AGR GENET BREEDING | 10 | 52% | 1.2% | 14 |
| 6 | SJTU BOR LUH FOOD SAFETY | 9 | 42% | 1.3% | 16 |
| 7 | REFERRAL MOL DIAG TRANSGEN PLANTING MAT | 8 | 100% | 0.4% | 5 |
| 8 | MOL BIOL GENOM UNIT | 8 | 42% | 1.2% | 14 |
| 9 | AGROBIOTECH | 7 | 42% | 1.1% | 13 |
| 10 | DETECTING AGR GENETICALLY MODIFIED ORGANISM | 6 | 80% | 0.3% | 4 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000150481 | SUBSTANTIAL EQUIVALENCE//PROTEIN ALLERGENICITY//GENETICALLY MODIFIED FEEDS |
| 2 | 0.0000120182 | AMANDIN//HIDDEN ALLERGENS//ALLERGEN DETECTION |
| 3 | 0.0000071211 | LOOP MEDIATED ISOTHERMAL AMPLIFICATION//LAMP//RT LAMP |
| 4 | 0.0000068211 | SPECIES IDENTIFICATION//MEAT SPECIES//FISH SPECIES IDENTIFICATION |
| 5 | 0.0000063302 | SERV BIOSANITARIOS//SOYBEAN PROTEINS//HEAT PROCESSED MEAT PRODUCTS |
| 6 | 0.0000061831 | DIGITAL PCR//DROPLET DIGITAL PCR//SINGLE CELL TRANSCRIPTOMICS |
| 7 | 0.0000057484 | RNA ISOLATION//RNA EXTRACTION//DNA ISOLATION |
| 8 | 0.0000053919 | CROP WILD HYBRIDIZATION//GENE ESCAPE//FARM SCALE EVALUATIONS |
| 9 | 0.0000052141 | OLEA EUROPAEA//OLEA EUROPAEA L//TOBACCO EXPERTISE |
| 10 | 0.0000051570 | GENOME WALKING//FALSE POSITIVE EXCLUSION//UNKNOWN FLANKING SEQUENCES |