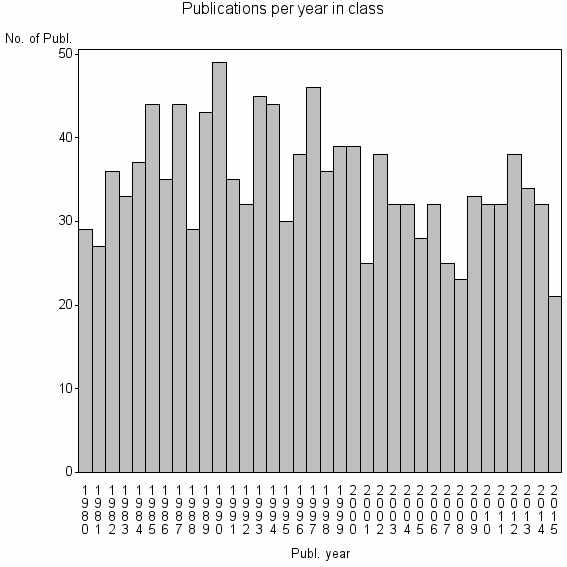
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 7814 | 1247 | 43.8 | 70% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 1083 | 9405 | ELECTROCHEMOTHERAPY//ELECTROPERMEABILIZATION//ELECTROPORATION |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | AMINOPHOSPHOLIPID TRANSLOCASE | Author keyword | 82 | 78% | 4% | 54 |
| 2 | PHOSPHOLIPID ASYMMETRY | Author keyword | 30 | 56% | 3% | 37 |
| 3 | FLIPPASE | Author keyword | 26 | 44% | 4% | 44 |
| 4 | LIPID ASYMMETRY | Author keyword | 24 | 55% | 2% | 30 |
| 5 | SCOTT SYNDROME | Author keyword | 19 | 74% | 1% | 14 |
| 6 | P4 ATPASE | Author keyword | 15 | 77% | 1% | 10 |
| 7 | SCRAMBLASE | Author keyword | 14 | 49% | 2% | 21 |
| 8 | PLSCR1 | Author keyword | 13 | 80% | 1% | 8 |
| 9 | PHOSPHOLIPID FLIPPASE | Author keyword | 12 | 86% | 0% | 6 |
| 10 | PHOSPHOLIPID SCRAMBLASE | Author keyword | 9 | 67% | 1% | 8 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | AMINOPHOSPHOLIPID TRANSLOCASE | 82 | 78% | 4% | 54 | Search AMINOPHOSPHOLIPID+TRANSLOCASE | Search AMINOPHOSPHOLIPID+TRANSLOCASE |
| 2 | PHOSPHOLIPID ASYMMETRY | 30 | 56% | 3% | 37 | Search PHOSPHOLIPID+ASYMMETRY | Search PHOSPHOLIPID+ASYMMETRY |
| 3 | FLIPPASE | 26 | 44% | 4% | 44 | Search FLIPPASE | Search FLIPPASE |
| 4 | LIPID ASYMMETRY | 24 | 55% | 2% | 30 | Search LIPID+ASYMMETRY | Search LIPID+ASYMMETRY |
| 5 | SCOTT SYNDROME | 19 | 74% | 1% | 14 | Search SCOTT+SYNDROME | Search SCOTT+SYNDROME |
| 6 | P4 ATPASE | 15 | 77% | 1% | 10 | Search P4+ATPASE | Search P4+ATPASE |
| 7 | SCRAMBLASE | 14 | 49% | 2% | 21 | Search SCRAMBLASE | Search SCRAMBLASE |
| 8 | PLSCR1 | 13 | 80% | 1% | 8 | Search PLSCR1 | Search PLSCR1 |
| 9 | PHOSPHOLIPID FLIPPASE | 12 | 86% | 0% | 6 | Search PHOSPHOLIPID+FLIPPASE | Search PHOSPHOLIPID+FLIPPASE |
| 10 | PHOSPHOLIPID SCRAMBLASE | 9 | 67% | 1% | 8 | Search PHOSPHOLIPID+SCRAMBLASE | Search PHOSPHOLIPID+SCRAMBLASE |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | TRANSBILAYER MOVEMENT | 128 | 48% | 16% | 197 |
| 2 | AMINOPHOSPHOLIPID TRANSLOCASE | 119 | 63% | 9% | 118 |
| 3 | SPIN LABELED PHOSPHOLIPIDS | 81 | 73% | 5% | 62 |
| 4 | LIPID ASYMMETRY | 77 | 61% | 7% | 83 |
| 5 | PHOSPHOLIPID TRANSLOCATION | 52 | 71% | 3% | 42 |
| 6 | PUTATIVE AMINOPHOSPHOLIPID TRANSLOCASES | 48 | 100% | 1% | 17 |
| 7 | PHOSPHATIDYLSERINE ASYMMETRY | 34 | 93% | 1% | 13 |
| 8 | DRS2P | 27 | 92% | 1% | 11 |
| 9 | AMINOPHOSPHOLIPID TRANSLOCASE ACTIVITY | 25 | 77% | 1% | 17 |
| 10 | TRANSMEMBRANE LIPID ASYMMETRY | 23 | 100% | 1% | 10 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Role of Flippases, Scramblases and Transfer Proteins in Phosphatidylserine Subcellular Distribution | 2015 | 6 | 96 | 49% |
| Pathophysiologic implications of membrane phospholipid asymmetry in blood cells | 1997 | 845 | 186 | 48% |
| Phospholipid flippases: Building asymmetric membranes and transport vesicles | 2012 | 44 | 110 | 63% |
| Lipid translocation across the plasma membrane of mammalian cells | 1999 | 270 | 108 | 73% |
| PHOSPHOLIPIDS IN ANIMAL EUKARYOTIC MEMBRANES - TRANSVERSE ASYMMETRY AND MOVEMENT | 1993 | 462 | 158 | 64% |
| Phospholipid scramblase: An update | 2010 | 90 | 62 | 58% |
| Regulation of transbilayer plasma membrane phospholipid asymmetry | 2003 | 225 | 142 | 63% |
| The Distribution and Function of Phosphatidylserine in Cellular Membranes | 2010 | 179 | 96 | 30% |
| Phospholipid flippases | 2007 | 105 | 65 | 57% |
| Chemical control of phospholipid distribution across bilayer membranes | 2002 | 125 | 182 | 67% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | MOL INTERACTKITA KU | 8 | 75% | 0.5% | 6 |
| 2 | MEMBRANE ENZYMOL | 5 | 28% | 1.2% | 15 |
| 3 | MOL INTERACT | 4 | 33% | 0.8% | 10 |
| 4 | PL IND MICROBIOL | 4 | 36% | 0.6% | 8 |
| 5 | BIOMEMBRANES MESSAGERS CELLULAI | 2 | 44% | 0.3% | 4 |
| 6 | MATH NAT WISSEN FAK 1 | 2 | 19% | 0.6% | 8 |
| 7 | MEMBRANE PUMPS CELLS DIS PUMPKIN | 1 | 11% | 0.8% | 10 |
| 8 | MACULAR | 1 | 15% | 0.6% | 7 |
| 9 | BIJVOET BIOMEMBRANES | 1 | 50% | 0.1% | 1 |
| 10 | BIOCHEM CELL BIOLSIBS | 1 | 50% | 0.1% | 1 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000150032 | PLATELET APOPTOSIS//COATED PLATELET//STUDY HUMAN OPERATOR PERFORMANCE |
| 2 | 0.0000132933 | STARA ZAGORA MED//THERMOHAEMOLYSIS//PHYS BIOPHYS ROENTGENOL RADIOL |
| 3 | 0.0000101274 | STEROL CARRIER PROTEIN 2//PHOSPHATIDYLINOSITOL TRANSFER PROTEIN//GLYCOLIPID TRANSFER PROTEIN |
| 4 | 0.0000097872 | ARTIFICIAL BOUNDARY LIPID//PROTEIN TRANSFER//GENOME BIOINFORMAT PROGRAM |
| 5 | 0.0000097330 | TC 99M ANNEXIN V//ANNEXIN V//APOPTOSIS IMAGING |
| 6 | 0.0000087934 | MEROCYANINE 540//BIOMED SPECT//MEROCYANINE 540 MC540 |
| 7 | 0.0000087612 | MFG E8//APOPTOTIC CELLS//CELL CLEARANCE |
| 8 | 0.0000076269 | ERYPTOSIS//CELL VOLUME//APOPTOTIC VOLUME DECREASE |
| 9 | 0.0000071846 | MICROPARTICLES//ENDOTHELIAL MICROPARTICLES//PLATELET DERIVED MICROPARTICLES |
| 10 | 0.0000070868 | SENESCENT CELL ANTIGEN//SENESCENT ERYTHROCYTES//ERYTHROCYTE AGING |