Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
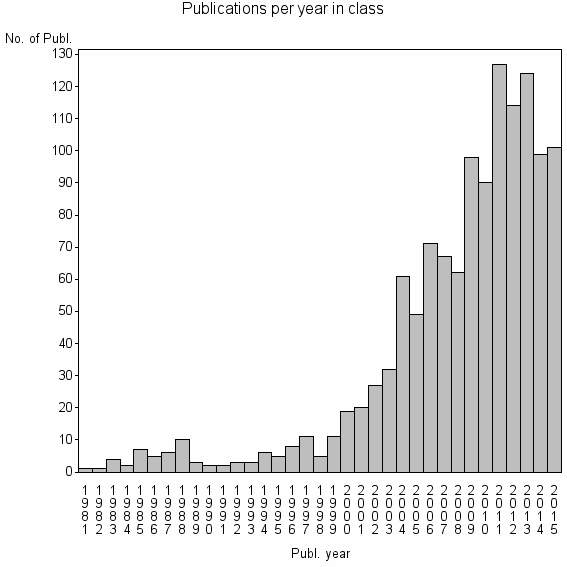
| 7735 | 1256 | 56.0 | 84% |
Classes in level above (level 2) |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | PLANT PROTEOMICS | Author keyword | 43 | 33% | 9% | 108 |
| 2 | INPPO | Author keyword | 12 | 86% | 0% | 6 |
| 3 | LEAF PROTEOME | Author keyword | 10 | 61% | 1% | 11 |
| 4 | ORG EDUC INITIAT | Address | 6 | 44% | 1% | 11 |
| 5 | PLANT PROTEIN EXTRACTION | Author keyword | 6 | 71% | 0% | 5 |
| 6 | AGR PLANT BIOCHEM PROTE GRP | Address | 5 | 45% | 1% | 9 |
| 7 | 2 DGE | Author keyword | 4 | 47% | 1% | 7 |
| 8 | PROTEIN EXTRACTION | Author keyword | 4 | 12% | 3% | 34 |
| 9 | BITCET | Address | 3 | 100% | 0% | 3 |
| 10 | CNRS UCBL INSA BAYER CROPSCI JOINT | Address | 3 | 100% | 0% | 3 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | PLANT PROTEOMICS | 43 | 33% | 9% | 108 | Search PLANT+PROTEOMICS | Search PLANT+PROTEOMICS |
| 2 | INPPO | 12 | 86% | 0% | 6 | Search INPPO | Search INPPO |
| 3 | LEAF PROTEOME | 10 | 61% | 1% | 11 | Search LEAF+PROTEOME | Search LEAF+PROTEOME |
| 4 | PLANT PROTEIN EXTRACTION | 6 | 71% | 0% | 5 | Search PLANT+PROTEIN+EXTRACTION | Search PLANT+PROTEIN+EXTRACTION |
| 5 | 2 DGE | 4 | 47% | 1% | 7 | Search 2+DGE | Search 2+DGE |
| 6 | PROTEIN EXTRACTION | 4 | 12% | 3% | 34 | Search PROTEIN+EXTRACTION | Search PROTEIN+EXTRACTION |
| 7 | DICOTYLEDONEOUS PLANTS | 3 | 100% | 0% | 3 | Search DICOTYLEDONEOUS+PLANTS | Search DICOTYLEDONEOUS+PLANTS |
| 8 | SALT RESPONSIVE PROTEINS | 3 | 100% | 0% | 3 | Search SALT+RESPONSIVE+PROTEINS | Search SALT+RESPONSIVE+PROTEINS |
| 9 | LEAF PROTEOMICS | 3 | 60% | 0% | 3 | Search LEAF+PROTEOMICS | Search LEAF+PROTEOMICS |
| 10 | PLANT PROTEOME | 2 | 44% | 0% | 4 | Search PLANT+PROTEOME | Search PLANT+PROTEOME |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | RECALCITRANT PLANT TISSUES | 65 | 71% | 4% | 52 |
| 2 | RESPONSIVE PROTEINS | 39 | 48% | 5% | 59 |
| 3 | DATA FILE | 31 | 92% | 1% | 12 |
| 4 | RICE LEAF SHEATH | 23 | 76% | 1% | 16 |
| 5 | POLYETHYLENE GLYCOL FRACTIONATION | 21 | 73% | 1% | 16 |
| 6 | SAMPLE EXTRACTION TECHNIQUES | 21 | 85% | 1% | 11 |
| 7 | 2 DE | 16 | 61% | 1% | 17 |
| 8 | PLANT PROTEOMICS | 13 | 58% | 1% | 15 |
| 9 | STRESS RESPONSIVE PROTEINS | 10 | 47% | 1% | 16 |
| 10 | TWO DIMENSIONAL ELECTROPHORESIS | 10 | 17% | 4% | 55 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Plant proteome changes under abiotic stress - Contribution of proteomics studies to understanding plant stress response | 2011 | 143 | 115 | 52% |
| Fourteen years of plant proteomics reflected in Proteomics: Moving from model species and 2DE-based approaches to orphan species and gel-free platforms | 2015 | 5 | 221 | 29% |
| Rice proteomics: A model system for crop improvement and food security | 2014 | 11 | 95 | 49% |
| Abiotic stress responses in plant roots: a proteomics perspective | 2014 | 14 | 126 | 35% |
| Mechanisms of Plant Salt Response: Insights from Proteomics | 2012 | 61 | 150 | 33% |
| Proteomics of stress responses in wheat and barley-search for potential protein markers of stress tolerance | 2014 | 7 | 72 | 49% |
| Protein Contribution to Plant Salinity Response and Tolerance Acquisition | 2013 | 15 | 103 | 47% |
| Uncovering plant-pathogen crosstalk through apoplastic proteomic studies | 2014 | 9 | 143 | 27% |
| Plant proteome responses to salinity stress - comparison of glycophytes and halophytes | 2013 | 9 | 80 | 59% |
| Rice proteomics: A move toward expanded proteome coverage to comparative and functional proteomics uncovers the mysteries of rice and plant biology | 2011 | 32 | 139 | 40% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | ORG EDUC INITIAT | 6 | 44% | 0.9% | 11 |
| 2 | AGR PLANT BIOCHEM PROTE GRP | 5 | 45% | 0.7% | 9 |
| 3 | BITCET | 3 | 100% | 0.2% | 3 |
| 4 | CNRS UCBL INSA BAYER CROPSCI JOINT | 3 | 100% | 0.2% | 3 |
| 5 | AGR PLANT BIOCHEM GRP | 3 | 50% | 0.3% | 4 |
| 6 | FOOD BIOCHEM ENGN | 3 | 50% | 0.3% | 4 |
| 7 | GRP PROTE GENOM FUNC PLANTAS | 3 | 60% | 0.2% | 3 |
| 8 | PROT METABOLITE DYNAM | 3 | 60% | 0.2% | 3 |
| 9 | ANIM PLANT INTRO | 2 | 67% | 0.2% | 2 |
| 10 | BREEDING OURCE DEV | 2 | 67% | 0.2% | 2 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000118943 | SAMPLE PREFRACTIONATION//PREFRACTIONATION//ALKALINE PROTEINS |
| 2 | 0.0000091382 | PLANT MICROBIAL GENOM//LRT1//NONADDITIVE PROTEIN |
| 3 | 0.0000088828 | ADV PLANT PHYSIOL//EPITHEM//SQUASH CUCURBITA |
| 4 | 0.0000074486 | HEAT STRESS TRANSCRIPTION FACTOR//THERMOTOLERANCE//HEAT SHOCK TRANSCRIPTION FACTOR |
| 5 | 0.0000070934 | FRUIT TREE SPECIES//ACID METABOLISM//SUCROSE COMPARTMENTATION |
| 6 | 0.0000070033 | DREB//AP2 ERF//GCC BOX |
| 7 | 0.0000069437 | WATER SOLUBLE CHLOROPHYLL BINDING PROTEIN//DUF538//CLASS II WSCP |
| 8 | 0.0000066183 | DEHYDRIN//LEA PROTEIN//DEHYDRINS |
| 9 | 0.0000060686 | RICE GENOME PROGRAM//CROP DESIGN//GENOME IL |
| 10 | 0.0000056278 | TWO DIMENSIONAL POLYACRYLAMIDE GEL ELECTROPHORESIS//MYOCARDIAL PROTEINS//PROTEOME GENE PROD M PING |