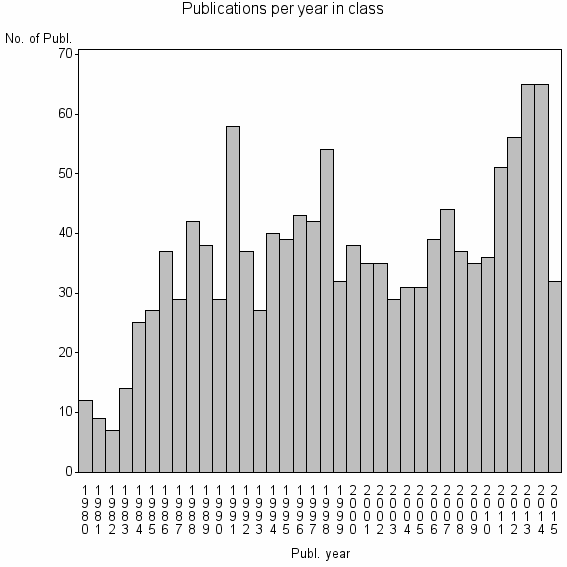
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 7339 | 1300 | 45.9 | 85% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 195 | 20594 | PRE MRNA SPLICING//RNA LOCALIZATION//SPLICEOSOME |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | POLYADENYLATION | Author keyword | 45 | 27% | 11% | 141 |
| 2 | ALTERNATIVE POLYADENYLATION | Author keyword | 30 | 43% | 4% | 53 |
| 3 | POLYA POLYMERASE | Author keyword | 25 | 44% | 3% | 43 |
| 4 | POLYA SITE | Author keyword | 23 | 86% | 1% | 12 |
| 5 | MRNA 3 END PROCESSING | Author keyword | 21 | 90% | 1% | 9 |
| 6 | PRE MRNA 3 END PROCESSING | Author keyword | 21 | 85% | 1% | 11 |
| 7 | CLEAVAGE AND POLYADENYLATION SPECIFICITY FACTOR | Author keyword | 14 | 100% | 1% | 7 |
| 8 | CLEAVAGE STIMULATION FACTOR | Author keyword | 11 | 100% | 0% | 6 |
| 9 | CLEAVAGE AND POLYADENYLATION | Author keyword | 11 | 69% | 1% | 9 |
| 10 | CLEAVAGE POLYADENYLATION | Author keyword | 9 | 83% | 0% | 5 |
Web of Science journal categories |
Author Key Words |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | POLYA SITE | 81 | 63% | 6% | 81 |
| 2 | MESSENGER RNA POLYADENYLATION | 77 | 52% | 8% | 105 |
| 3 | AAUAAA | 74 | 79% | 4% | 48 |
| 4 | POLYADENYLATION SPECIFICITY FACTOR | 71 | 71% | 4% | 57 |
| 5 | ALTERNATIVE POLYADENYLATION | 69 | 46% | 9% | 111 |
| 6 | MAMMALIAN CLEAVAGE | 69 | 93% | 2% | 26 |
| 7 | 3 END FORMATION | 68 | 33% | 13% | 173 |
| 8 | CLEAVAGE FACTOR I | 50 | 86% | 2% | 25 |
| 9 | SPECIFICITY FACTOR | 45 | 40% | 7% | 88 |
| 10 | CLEAVAGE POLYADENYLATION | 38 | 89% | 1% | 17 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Alternative mRNA transcription, processing, and translation: insights from RNA sequencing | 2015 | 6 | 139 | 29% |
| Alternative cleavage and polyadenylation: extent, regulation and function | 2013 | 75 | 85 | 65% |
| Ending the message: poly(A) signals then and now | 2011 | 146 | 116 | 63% |
| Alternative cleavage and polyadenylation: the long and short of it | 2013 | 52 | 103 | 65% |
| Mechanism and regulation of mRNA polyadenylation | 1997 | 457 | 85 | 76% |
| Formation of mRNA 3 ' ends in eukaryotes: Mechanism, regulation, and interrelationships with other steps in mRNA synthesis | 1999 | 676 | 481 | 47% |
| Mechanisms and Consequences of Alternative Polyadenylation | 2011 | 163 | 133 | 40% |
| Protein factors in pre-mRNA 3'-end processing | 2008 | 184 | 219 | 62% |
| Delineating the Structural Blueprint of the Pre-mRNA 3 '-End Processing Machinery | 2014 | 5 | 192 | 73% |
| THE BIOCHEMISTRY OF 3'-END CLEAVAGE AND POLYADENYLATION OF MESSENGER-RNA PRECURSORS | 1992 | 345 | 165 | 70% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | MINIST EDUC COSTAL WETLAND ECOSYST | 6 | 100% | 0.3% | 4 |
| 2 | P ANIKOLAOU | 5 | 60% | 0.5% | 6 |
| 3 | P ANIKOLAOU ONCOL | 3 | 57% | 0.3% | 4 |
| 4 | STATE BIOCONTROLGUANGDONG PROV P | 3 | 60% | 0.2% | 3 |
| 5 | INSERM ERM 206 | 1 | 100% | 0.2% | 2 |
| 6 | CELL ACTIVAT GENE EXP S GRP | 1 | 40% | 0.2% | 2 |
| 7 | CELL VIRUS GENET GRP | 1 | 50% | 0.1% | 1 |
| 8 | DIGNA BIOTECH | 1 | 50% | 0.1% | 1 |
| 9 | ELECT MATH SCI ENGN | 1 | 50% | 0.1% | 1 |
| 10 | GENET GEN DEV | 1 | 50% | 0.1% | 1 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000162878 | MRNP BIOGENESIS METAB//DIS3//RRP6 |
| 2 | 0.0000153152 | CYTOPLASMIC POLYADENYLATION//PUMILIO//CPEB |
| 3 | 0.0000125168 | P TEFB//CDK9//HEXIM1 |
| 4 | 0.0000112515 | POLYA BINDING PROTEIN//POLY A BINDING PROTEIN//PABP |
| 5 | 0.0000111593 | OCULOPHARYNGEAL MUSCULAR DYSTROPHY//PABPN1//OPMD |
| 6 | 0.0000100606 | IGD MYELOMA//IGD RECEPTOR//IMMUNOGLOBULIN D |
| 7 | 0.0000100322 | TRNA NUCLEOTIDYLTRANSFERASE//CCA ADDING ENZYME//TRNAHIS GUANYLYLTRANSFERASE |
| 8 | 0.0000080314 | HNRNP B1//HETEROGENEOUS NUCLEAR RIBONUCLEOPROTEINS HNRNPS//RNP CONCENSUS RNA BINDING DOMAIN |
| 9 | 0.0000071138 | HISTONE GENE EXPRESSION//HISTONE MRNA//SLBP |
| 10 | 0.0000060714 | ANTI THY 12 ANTIBODY//CELL TYPE IDENTITY//ENHANCER CORE |