Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
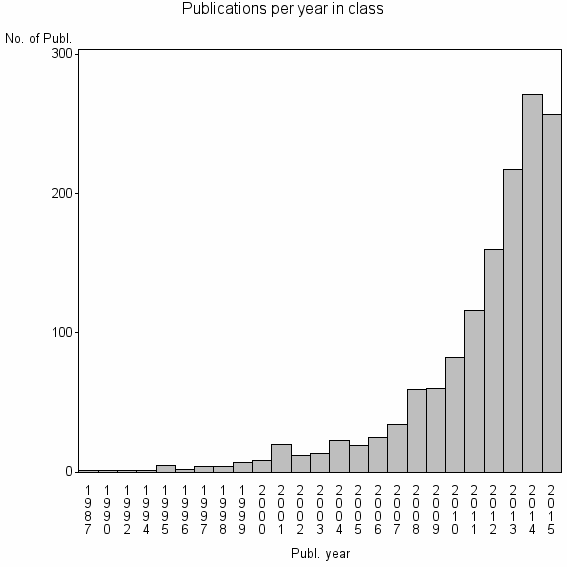
| 6552 | 1402 | 37.3 | 92% |
Classes in level above (level 2) |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | ROLLING CIRCLE AMPLIFICATION | Author keyword | 24 | 33% | 4% | 60 |
| 2 | HYBRIDIZATION CHAIN REACTION | Author keyword | 23 | 52% | 2% | 31 |
| 3 | DNAZYME | Author keyword | 20 | 25% | 5% | 72 |
| 4 | PADLOCK PROBES | Author keyword | 19 | 68% | 1% | 17 |
| 5 | MICRORNA DETECTION | Author keyword | 11 | 69% | 1% | 9 |
| 6 | TERMINAL PROTECTION | Author keyword | 10 | 73% | 1% | 8 |
| 7 | SINGLE MOL DETECT IMAGING | Address | 9 | 39% | 1% | 19 |
| 8 | CLIN MOL MED DETECT | Address | 9 | 83% | 0% | 5 |
| 9 | IMMUNO RCA | Author keyword | 9 | 83% | 0% | 5 |
| 10 | EXONUCLEASE III | Author keyword | 9 | 32% | 2% | 23 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | ROLLING CIRCLE AMPLIFICATION | 24 | 33% | 4% | 60 | Search ROLLING+CIRCLE+AMPLIFICATION | Search ROLLING+CIRCLE+AMPLIFICATION |
| 2 | HYBRIDIZATION CHAIN REACTION | 23 | 52% | 2% | 31 | Search HYBRIDIZATION+CHAIN+REACTION | Search HYBRIDIZATION+CHAIN+REACTION |
| 3 | DNAZYME | 20 | 25% | 5% | 72 | Search DNAZYME | Search DNAZYME |
| 4 | PADLOCK PROBES | 19 | 68% | 1% | 17 | Search PADLOCK+PROBES | Search PADLOCK+PROBES |
| 5 | MICRORNA DETECTION | 11 | 69% | 1% | 9 | Search MICRORNA+DETECTION | Search MICRORNA+DETECTION |
| 6 | TERMINAL PROTECTION | 10 | 73% | 1% | 8 | Search TERMINAL+PROTECTION | Search TERMINAL+PROTECTION |
| 7 | IMMUNO RCA | 9 | 83% | 0% | 5 | Search IMMUNO+RCA | Search IMMUNO+RCA |
| 8 | EXONUCLEASE III | 9 | 32% | 2% | 23 | Search EXONUCLEASE+III | Search EXONUCLEASE+III |
| 9 | G QUADRUPLEX DNAZYME | 8 | 100% | 0% | 5 | Search G+QUADRUPLEX+DNAZYME | Search G+QUADRUPLEX+DNAZYME |
| 10 | PADLOCK PROBE | 7 | 46% | 1% | 12 | Search PADLOCK+PROBE | Search PADLOCK+PROBE |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | PADLOCK PROBES | 66 | 51% | 6% | 91 |
| 2 | HYBRIDIZATION CHAIN REACTION | 63 | 58% | 5% | 72 |
| 3 | ROLLING CIRCLE AMPLIFICATION | 62 | 27% | 14% | 201 |
| 4 | NICKING ENDONUCLEASE | 40 | 82% | 2% | 23 |
| 5 | SIGNAL AMPLIFICATION | 38 | 16% | 15% | 213 |
| 6 | ISOTHERMAL AMPLIFICATION | 38 | 51% | 4% | 52 |
| 7 | AMPLIFIED DETECTION | 25 | 38% | 4% | 53 |
| 8 | ENZYME FREE | 24 | 54% | 2% | 31 |
| 9 | RAMIFICATION AMPLIFICATION | 21 | 85% | 1% | 11 |
| 10 | DNAZYME | 21 | 22% | 6% | 82 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Rolling circle amplification: a versatile tool for chemical biology, materials science and medicine | 2014 | 37 | 147 | 53% |
| The hybridization chain reaction in the development of ultrasensitive nucleic acid assays | 2015 | 3 | 65 | 49% |
| A review: microRNA detection methods | 2015 | 2 | 64 | 66% |
| MicroRNA profiling: approaches and considerations | 2012 | 245 | 107 | 20% |
| Enzyme-assisted target recycling (EATR) for nucleic acid detection | 2014 | 20 | 179 | 46% |
| Peroxidase-mimicking DNAzymes for biosensing applications: A review | 2011 | 61 | 69 | 61% |
| Isothermal amplified detection of DNA and RNA | 2014 | 22 | 247 | 35% |
| DNA-Mediated Homogeneous Binding Assays for Nucleic Acids and Proteins | 2013 | 63 | 302 | 27% |
| MicroRNA: Function, Detection, and Bioanalysis | 2013 | 77 | 323 | 23% |
| Rolling circle amplification: Applications in nanotechnology and biodetection with functional nucleic acids | 2008 | 157 | 90 | 41% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | SINGLE MOL DETECT IMAGING | 9 | 39% | 1.4% | 19 |
| 2 | CLIN MOL MED DETECT | 9 | 83% | 0.4% | 5 |
| 3 | JIANGSU TARGET DRUG CLIN PLICAT | 4 | 75% | 0.2% | 3 |
| 4 | MOE BIOORGAN CHEM MOL ENGN | 4 | 29% | 0.8% | 11 |
| 5 | MINERVA COMPLEX BIOHYBRID SYST | 3 | 100% | 0.2% | 3 |
| 6 | SENSOR ANAL TUMOR MARKER | 3 | 19% | 0.9% | 13 |
| 7 | BIOSYST MICROANAL | 2 | 17% | 0.9% | 13 |
| 8 | LUMINESCENT REAL TIME ANALYT CHEM | 2 | 13% | 1.1% | 16 |
| 9 | BIOCHEM ANAL | 2 | 13% | 0.9% | 13 |
| 10 | NAT PROD CHEM BIOL | 2 | 43% | 0.2% | 3 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000216515 | DNAZYME//DEOXYRIBOZYME//CATALYTIC DNA |
| 2 | 0.0000139698 | IMMUNO PCR//LEHRSTUHL BIOL CHEM MIKROSTRUKTURTECH//REAL TIME IMMUNO PCR |
| 3 | 0.0000129602 | APTAMER//SELEX//APTAMERS |
| 4 | 0.0000102690 | COLORIMETRIC DETECTION//MERCURY ION//MERCURY IONS |
| 5 | 0.0000093789 | DNA ORIGAMI//DNA NANOTECHNOLOGY//DNA NANOSTRUCTURES |
| 6 | 0.0000093731 | LOOP MEDIATED ISOTHERMAL AMPLIFICATION//LAMP//RT LAMP |
| 7 | 0.0000093591 | INT NANOTECHNOL//CIGMH//DNA MATERIALS |
| 8 | 0.0000093002 | ELECTROCHEMICAL DNA BIOSENSOR//DNA BIOSENSOR//GENOSENSOR |
| 9 | 0.0000080451 | NEWCAT//MOLECULAR BEACON//MOLECULAR BEACONS |
| 10 | 0.0000079265 | SURFACE MEDIATED POLYMERIZATION//VISIBLE LIGHT PHOTOPOLYMERIZATION//ACCESSIBILITY PREDICTION |