Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
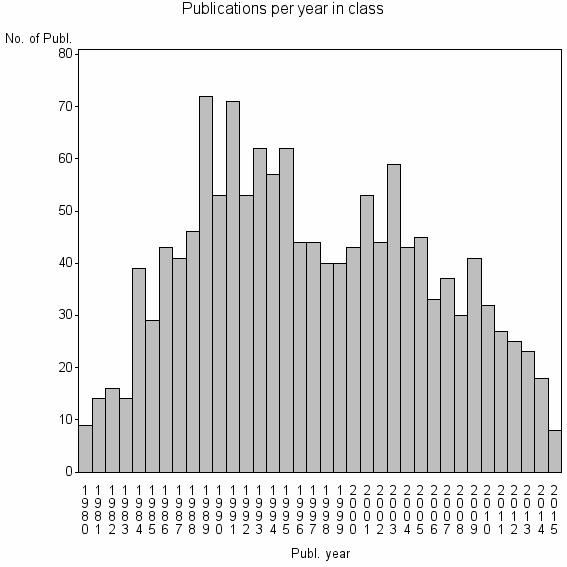
| 6481 | 1410 | 40.3 | 71% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 777 | 11593 | MADS BOX GENES//MADS BOX//FLOWER DEVELOPMENT |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | AC DS | Author keyword | 47 | 79% | 2% | 30 |
| 2 | TRANSPOSON TAGGING | Author keyword | 44 | 59% | 3% | 49 |
| 3 | AC TRANSPOSASE | Author keyword | 21 | 90% | 1% | 9 |
| 4 | AC TRANSPOSON | Author keyword | 15 | 88% | 0% | 7 |
| 5 | ACTIVATION TAGGING | Author keyword | 12 | 34% | 2% | 29 |
| 6 | GENE TAGGING | Author keyword | 12 | 31% | 2% | 32 |
| 7 | T DNA | Author keyword | 11 | 21% | 3% | 48 |
| 8 | DS ELEMENT | Author keyword | 11 | 100% | 0% | 6 |
| 9 | EN SPM | Author keyword | 10 | 73% | 1% | 8 |
| 10 | DTPH1 | Author keyword | 9 | 83% | 0% | 5 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | AC DS | 47 | 79% | 2% | 30 | Search AC+DS | Search AC+DS |
| 2 | TRANSPOSON TAGGING | 44 | 59% | 3% | 49 | Search TRANSPOSON+TAGGING | Search TRANSPOSON+TAGGING |
| 3 | AC TRANSPOSASE | 21 | 90% | 1% | 9 | Search AC+TRANSPOSASE | Search AC+TRANSPOSASE |
| 4 | AC TRANSPOSON | 15 | 88% | 0% | 7 | Search AC+TRANSPOSON | Search AC+TRANSPOSON |
| 5 | ACTIVATION TAGGING | 12 | 34% | 2% | 29 | Search ACTIVATION+TAGGING | Search ACTIVATION+TAGGING |
| 6 | GENE TAGGING | 12 | 31% | 2% | 32 | Search GENE+TAGGING | Search GENE+TAGGING |
| 7 | T DNA | 11 | 21% | 3% | 48 | Search T+DNA | Search T+DNA |
| 8 | DS ELEMENT | 11 | 100% | 0% | 6 | Search DS+ELEMENT | Search DS+ELEMENT |
| 9 | EN SPM | 10 | 73% | 1% | 8 | Search EN+SPM | Search EN+SPM |
| 10 | DTPH1 | 9 | 83% | 0% | 5 | Search DTPH1 | Search DTPH1 |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | ACTIVATOR AC | 141 | 93% | 4% | 53 |
| 2 | ELEMENT ACTIVATOR AC | 65 | 93% | 2% | 25 |
| 3 | DISSOCIATION EXCISION | 53 | 89% | 2% | 24 |
| 4 | AC TRANSPOSITION | 47 | 81% | 2% | 29 |
| 5 | ELEMENT SYSTEM | 45 | 83% | 2% | 25 |
| 6 | SOMATIC MUTABILITY | 41 | 100% | 1% | 15 |
| 7 | ELEMENT AC | 38 | 86% | 1% | 19 |
| 8 | DNA MODIFICATION | 36 | 56% | 3% | 45 |
| 9 | GERMINAL TRANSPOSITION | 34 | 93% | 1% | 13 |
| 10 | MAIZE ELEMENT ACTIVATOR | 32 | 85% | 1% | 17 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Contribution of the Tos17 retrotransposon to rice functional genomics | 2001 | 139 | 36 | 53% |
| Phenome Analysis in Plant Species Using Loss-of-Function and Gain-of-Function Mutants | 2009 | 31 | 108 | 56% |
| STRATEGIES FOR MUTAGENESIS AND GENE CLONING USING TRANSPOSON TAGGING AND T-DNA INSERTIONAL MUTAGENESIS | 1992 | 158 | 86 | 79% |
| Mutant Resources for the Functional Analysis of the Rice Genome | 2013 | 10 | 64 | 39% |
| Reverse genetic approaches for functional genomics of rice | 2005 | 52 | 113 | 50% |
| The art and design of genetic screens: maize | 2008 | 28 | 84 | 36% |
| Arabidopsis gene knockout: phenotypes wanted | 2001 | 154 | 48 | 29% |
| Mutator transposons | 2002 | 93 | 48 | 69% |
| The Mutator transposable element system of maize | 1996 | 66 | 110 | 78% |
| Molecular genetics using T-DNA in rice | 2005 | 52 | 115 | 41% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | RICE FUNCT GENOM GRP | 2 | 32% | 0.4% | 6 |
| 2 | ROY J CARVER CO 2035B | 2 | 67% | 0.1% | 2 |
| 3 | WARTIK 519 | 1 | 38% | 0.2% | 3 |
| 4 | DEV BIOL CHIKUSA KU | 1 | 50% | 0.1% | 2 |
| 5 | RICE FUNCT GENOM | 1 | 50% | 0.1% | 2 |
| 6 | SHANGHAI BIOENERGY CROP | 1 | 25% | 0.2% | 3 |
| 7 | CEREAL TRANSFORMAT GRP | 1 | 50% | 0.1% | 1 |
| 8 | CNRSUMR 5096 | 1 | 50% | 0.1% | 1 |
| 9 | DNA ANALYT GENOM | 1 | 50% | 0.1% | 1 |
| 10 | GENOMINE INC | 1 | 50% | 0.1% | 1 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000182905 | RETROTRANSPOSONS//TY1 COPIA//TRANSPOSABLE ELEMENTS |
| 2 | 0.0000155713 | RICE GENOME PROGRAM//CROP DESIGN//GENOME IL |
| 3 | 0.0000138834 | MARKER FREE TRANSGENIC PLANTS//MARKER FREE//MANNOSE SELECTION |
| 4 | 0.0000138302 | TILLING//SURVEYOR NUCLEASE//ECOTILLING |
| 5 | 0.0000092245 | CALIBRACHOA//PETUNIA INTEGRIFOLIA//PETUNIA |
| 6 | 0.0000088352 | GENOME WALKING//FALSE POSITIVE EXCLUSION//UNKNOWN FLANKING SEQUENCES |
| 7 | 0.0000075123 | ANTHOCYANIN//ANTHOCYANIN BIOSYNTHESIS//FLAVONOL SYNTHASE |
| 8 | 0.0000069495 | MARINER//SLEEPING BEAUTY//ARNOLD MABEL BECKMAN TRANSPOSON |
| 9 | 0.0000065161 | CROWN GALL//AGROBACTERIUM VITIS//TI PLASMID |
| 10 | 0.0000063766 | ENDOSPERM TRANSFER CELLS//EMBRYO SURROUNDING REGION//DEK1 |