Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
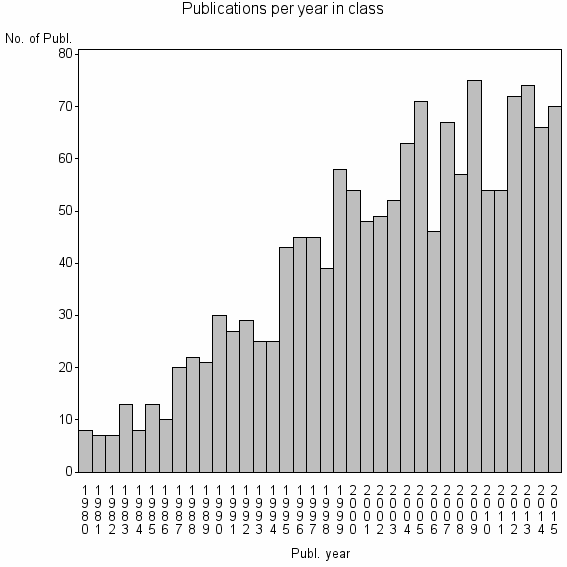
| 6073 | 1467 | 47.2 | 85% |
Classes in level above (level 2) |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | EIF4E | Author keyword | 119 | 51% | 11% | 166 |
| 2 | EIF 4E | Author keyword | 25 | 70% | 1% | 21 |
| 3 | CAPPING EFFICIENCY | Author keyword | 20 | 100% | 1% | 9 |
| 4 | CAP ANALOG | Author keyword | 15 | 82% | 1% | 9 |
| 5 | EIF4F | Author keyword | 15 | 50% | 1% | 22 |
| 6 | EIF4E PHOSPHORYLATION | Author keyword | 15 | 88% | 0% | 7 |
| 7 | MRNA 5 CAP | Author keyword | 14 | 100% | 0% | 7 |
| 8 | 4E BP1 | Author keyword | 13 | 27% | 3% | 42 |
| 9 | EUKARYOTIC INITIATION FACTOR 4E | Author keyword | 12 | 39% | 2% | 24 |
| 10 | MRNA CAP STRUCTURE | Author keyword | 12 | 86% | 0% | 6 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | EIF4E | 119 | 51% | 11% | 166 | Search EIF4E | Search EIF4E |
| 2 | EIF 4E | 25 | 70% | 1% | 21 | Search EIF+4E | Search EIF+4E |
| 3 | CAPPING EFFICIENCY | 20 | 100% | 1% | 9 | Search CAPPING+EFFICIENCY | Search CAPPING+EFFICIENCY |
| 4 | CAP ANALOG | 15 | 82% | 1% | 9 | Search CAP+ANALOG | Search CAP+ANALOG |
| 5 | EIF4F | 15 | 50% | 1% | 22 | Search EIF4F | Search EIF4F |
| 6 | EIF4E PHOSPHORYLATION | 15 | 88% | 0% | 7 | Search EIF4E+PHOSPHORYLATION | Search EIF4E+PHOSPHORYLATION |
| 7 | MRNA 5 CAP | 14 | 100% | 0% | 7 | Search MRNA+5++CAP | Search MRNA+5++CAP |
| 8 | 4E BP1 | 13 | 27% | 3% | 42 | Search 4E+BP1 | Search 4E+BP1 |
| 9 | EUKARYOTIC INITIATION FACTOR 4E | 12 | 39% | 2% | 24 | Search EUKARYOTIC+INITIATION+FACTOR+4E | Search EUKARYOTIC+INITIATION+FACTOR+4E |
| 10 | MRNA CAP STRUCTURE | 12 | 86% | 0% | 6 | Search MRNA+CAP+STRUCTURE | Search MRNA+CAP+STRUCTURE |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | FACTOR EIF 4E | 320 | 85% | 12% | 169 |
| 2 | INITIATION FACTOR 4E | 184 | 47% | 20% | 292 |
| 3 | MESSENGER RNA CAP | 152 | 74% | 8% | 113 |
| 4 | CAP BINDING PROTEIN | 132 | 45% | 15% | 218 |
| 5 | FACTOR 4E | 131 | 65% | 8% | 124 |
| 6 | EIF4E | 111 | 54% | 10% | 141 |
| 7 | TRANSLATION FACTOR EIF4E | 61 | 95% | 1% | 20 |
| 8 | EIF 4E | 53 | 79% | 2% | 34 |
| 9 | BINDING PROTEIN EIF4E | 41 | 90% | 1% | 18 |
| 10 | CAP DEPENDENT TRANSLATION | 41 | 35% | 7% | 96 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Targeting the eukaryotic translation initiation factor 4E for cancer therapy | 2008 | 161 | 19 | 89% |
| eIF4 initiation factors: Effectors of mRNA recruitment to ribosomes and regulators of translation | 1999 | 1267 | 341 | 39% |
| Targeting Mnks for Cancer Therapy | 2012 | 33 | 93 | 66% |
| Targeting the eIF4F Translation Initiation Complex: A Critical Nexus for Cancer Development | 2015 | 2 | 194 | 56% |
| MAP Kinase-Interacting Kinases-Emerging Targets against Cancer | 2014 | 9 | 61 | 74% |
| Targeting the translation machinery in cancer | 2015 | 3 | 272 | 39% |
| Translational control in cancer | 2010 | 238 | 139 | 28% |
| Regulation of Translation Initiation in Eukaryotes: Mechanisms and Biological Targets | 2009 | 818 | 128 | 11% |
| eIF-4E expression and its role in malignancies and metastases | 2004 | 394 | 122 | 48% |
| Cap and cap-binding proteins in the control of gene expression | 2011 | 71 | 228 | 38% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | POSTTRANSCRIPT CONTROL GRP | 7 | 48% | 0.7% | 11 |
| 2 | EQUIPE TRADUCT CYCLE CELLULAIRE DEV | 5 | 55% | 0.4% | 6 |
| 3 | MCGILL CANC | 4 | 10% | 2.8% | 41 |
| 4 | TRANSLAT CONTROL GRP | 4 | 46% | 0.4% | 6 |
| 5 | HEME ONC TRANSPLANT | 2 | 67% | 0.1% | 2 |
| 6 | UMR 7150EQUIPE CYCLE CELLULAIRE DEV | 2 | 67% | 0.1% | 2 |
| 7 | UMR 7150 | 2 | 22% | 0.5% | 7 |
| 8 | UMR MER SANTE 7150 | 2 | 43% | 0.2% | 3 |
| 9 | AMBION INC | 1 | 50% | 0.1% | 2 |
| 10 | CANC GROWTH TRANSLAT GENET | 1 | 50% | 0.1% | 2 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000187631 | IRES//INTERNAL RIBOSOME ENTRY SITE IRES//INTERNAL RIBOSOME ENTRY SITE |
| 2 | 0.0000180810 | EIF3//GENE REGULAT DEV//EIF1 |
| 3 | 0.0000134600 | POLYA BINDING PROTEIN//POLY A BINDING PROTEIN//PABP |
| 4 | 0.0000131834 | RNA HELICASE//DEAD BOX//DEAD BOX PROTEIN |
| 5 | 0.0000117369 | MTOR//RAPAMYCIN//CCI 779 |
| 6 | 0.0000106321 | MRNA DELIVERY//MRNA TRANSFECTION//RIBOL |
| 7 | 0.0000097705 | ARENE INTERACTIONS//U MOTIF//GUANINE BASE |
| 8 | 0.0000097172 | PDCD4//PROGRAMMED CELL DEATH 4//PROGRAMMED CELL DEATH 4 PDCD4 |
| 9 | 0.0000095504 | S6K2//S6 KINASE//RIBOSOMAL S6 KINASE 2 S6K2 |
| 10 | 0.0000082986 | DECAPPING//P BODIES//CCR4 NOT |