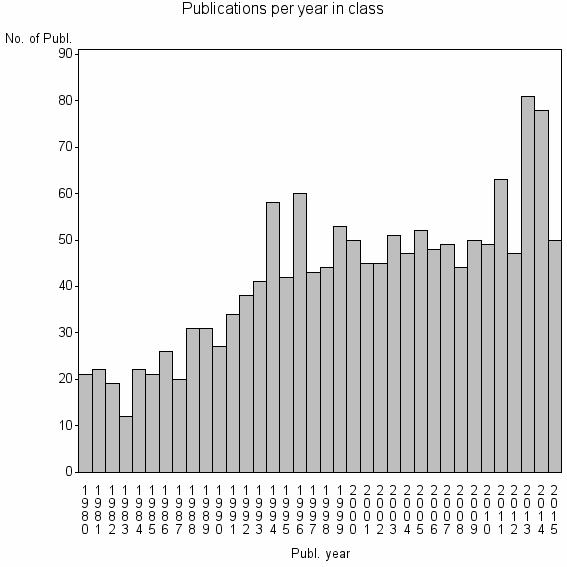
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 5748 | 1514 | 40.3 | 76% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 540 | 13989 | FLUOROQUINOLONES//CIPROFLOXACIN//QUINOLONES |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | DNA GYRASE | Author keyword | 55 | 41% | 7% | 104 |
| 2 | GYRASE | Author keyword | 32 | 49% | 3% | 47 |
| 3 | REVERSE GYRASE | Author keyword | 30 | 84% | 1% | 16 |
| 4 | CLOROBIOCIN | Author keyword | 15 | 82% | 1% | 9 |
| 5 | SUPERCOILING | Author keyword | 11 | 24% | 3% | 40 |
| 6 | DNA TOPOLOGY | Author keyword | 11 | 20% | 3% | 47 |
| 7 | NOVIOSE | Author keyword | 11 | 78% | 0% | 7 |
| 8 | POSITIVE SUPERCOILING | Author keyword | 11 | 78% | 0% | 7 |
| 9 | DNA SUPERCOILING | Author keyword | 9 | 21% | 3% | 38 |
| 10 | AMINOCOUMARIN ANTIBIOTICS | Author keyword | 9 | 83% | 0% | 5 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | DNA GYRASE | 55 | 41% | 7% | 104 | Search DNA+GYRASE | Search DNA+GYRASE |
| 2 | GYRASE | 32 | 49% | 3% | 47 | Search GYRASE | Search GYRASE |
| 3 | REVERSE GYRASE | 30 | 84% | 1% | 16 | Search REVERSE+GYRASE | Search REVERSE+GYRASE |
| 4 | CLOROBIOCIN | 15 | 82% | 1% | 9 | Search CLOROBIOCIN | Search CLOROBIOCIN |
| 5 | SUPERCOILING | 11 | 24% | 3% | 40 | Search SUPERCOILING | Search SUPERCOILING |
| 6 | DNA TOPOLOGY | 11 | 20% | 3% | 47 | Search DNA+TOPOLOGY | Search DNA+TOPOLOGY |
| 7 | NOVIOSE | 11 | 78% | 0% | 7 | Search NOVIOSE | Search NOVIOSE |
| 8 | POSITIVE SUPERCOILING | 11 | 78% | 0% | 7 | Search POSITIVE+SUPERCOILING | Search POSITIVE+SUPERCOILING |
| 9 | DNA SUPERCOILING | 9 | 21% | 3% | 38 | Search DNA+SUPERCOILING | Search DNA+SUPERCOILING |
| 10 | AMINOCOUMARIN ANTIBIOTICS | 9 | 83% | 0% | 5 | Search AMINOCOUMARIN+ANTIBIOTICS | Search AMINOCOUMARIN+ANTIBIOTICS |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | GYRASE GENES | 48 | 65% | 3% | 46 |
| 2 | N TERMINAL FRAGMENT | 45 | 36% | 7% | 100 |
| 3 | 2 GATE MECHANISM | 44 | 66% | 3% | 41 |
| 4 | B PROTEIN | 44 | 42% | 5% | 82 |
| 5 | STRAND PASSAGE | 44 | 72% | 2% | 34 |
| 6 | COUMERMYCIN A1 | 41 | 81% | 2% | 25 |
| 7 | LEU 500 PROMOTER | 41 | 81% | 2% | 25 |
| 8 | GYRASE | 40 | 27% | 8% | 124 |
| 9 | NOVOBIOCIN | 36 | 33% | 6% | 90 |
| 10 | IIA TOPOISOMERASES | 36 | 71% | 2% | 29 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Non-quinolone Inhibitors of Bacterial Type IIA Topoisomerases: A Feat of Bioisosterism | 2014 | 21 | 278 | 54% |
| DNA topoisomerases: harnessing and constraining energy to govern chromosome topology | 2008 | 166 | 274 | 49% |
| Exploiting bacterial DNA gyrase as a drug target: current state and perspectives | 2011 | 61 | 151 | 61% |
| Quinolone-mediated bacterial death | 2008 | 146 | 72 | 51% |
| All tangled up: how cells direct, manage and exploit topoisomerase function | 2011 | 98 | 199 | 34% |
| Prospects for Developing New Antibacterials Targeting Bacterial Type IIA Topoisomerases | 2014 | 7 | 135 | 61% |
| Structure, molecular mechanisms, and evolutionary relationships in DNA topoisomerases | 2004 | 246 | 155 | 47% |
| DNA gyrase, topoisomerase IV, and the 4-quinolones | 1997 | 693 | 174 | 39% |
| Thirty years of Escherichia coli DNA gyrase: From in vivo function to single-molecule mechanism | 2007 | 58 | 95 | 85% |
| Growth inhibition mediated by excess negative supercoiling: the interplay between transcription elongation, R-loop formation and DNA topology | 2006 | 79 | 64 | 56% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | ENZYMOL ACIDES NUCL | 7 | 64% | 0.5% | 7 |
| 2 | PHARMAZEUT BIOL PHARMAZEUT | 3 | 100% | 0.2% | 3 |
| 3 | INFECT DIS CEDD | 2 | 38% | 0.3% | 5 |
| 4 | SCI PL COMP | 2 | 67% | 0.1% | 2 |
| 5 | INFECT INNOVAT MED UNIT | 2 | 17% | 0.7% | 11 |
| 6 | INFECT IMED | 2 | 29% | 0.4% | 6 |
| 7 | MOL METAB SIGNALLING | 2 | 26% | 0.4% | 6 |
| 8 | DIS DEV WORLD CEDD | 1 | 100% | 0.1% | 2 |
| 9 | FRE 2445EQUIPE MICROBIOL | 1 | 100% | 0.1% | 2 |
| 10 | GENET MICROBIOL ENZYMOL ACIDES NUCL | 1 | 100% | 0.1% | 2 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000200716 | TOPOISOMERASE II//ICRF 193//DNA TOPOISOMERASE II |
| 2 | 0.0000114222 | QNR//QEPA//AAC6 IB CR |
| 3 | 0.0000100358 | GEMIFLOXACIN//GEMIFLOXACIN MESYLATE//GARENOXACIN |
| 4 | 0.0000096950 | DIHYDROPYRAZOLE//OXIME ESTER//ULCEROGENIC EFFECT |
| 5 | 0.0000093600 | H NS//HISTONE LIKE PROTEIN//BACTERIAL CHROMATIN |
| 6 | 0.0000084444 | N SUBSTITUTED PIPERAZINYL QUINOLONES//PIPERAZINYL QUINOLONES//S ENOKI 3394 |
| 7 | 0.0000072153 | EPIDEMIOLOGICAL SIGNIFICANCE//SIMILARITY IDENTIFICATION//AIRBORNE ESCHERICHIA COLI |
| 8 | 0.0000068832 | PARB//DNA SEGREGATION//FTSK |
| 9 | 0.0000065542 | ROPELENGTH//MOBIUS ENERGY//KNOT ENERGY |
| 10 | 0.0000064351 | C 14 PRECURSORS//CHEM TECHNOL TECH MICROBIOL BIOCHEM//AMMONIUM SALTS QUATERNARY |