Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
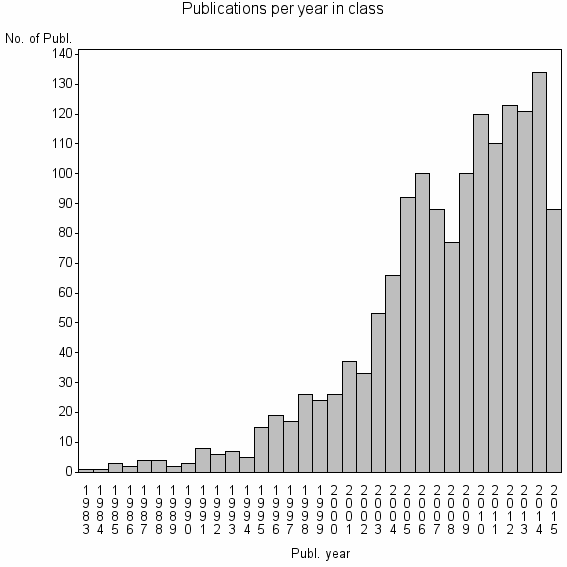
| 5743 | 1515 | 56.2 | 93% |
Classes in level above (level 2) |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | HISTONE CHAPERONE | Author keyword | 70 | 67% | 4% | 64 |
| 2 | ASF1 | Author keyword | 53 | 85% | 2% | 28 |
| 3 | CAF 1 | Author keyword | 32 | 78% | 1% | 21 |
| 4 | MACROH2A | Author keyword | 30 | 79% | 1% | 19 |
| 5 | CHROMATIN ASSEMBLY | Author keyword | 29 | 51% | 3% | 41 |
| 6 | HISTONE VARIANT | Author keyword | 25 | 47% | 3% | 40 |
| 7 | HISTONE VARIANTS | Author keyword | 25 | 44% | 3% | 43 |
| 8 | H2AZ | Author keyword | 25 | 49% | 2% | 37 |
| 9 | NUCLEOSOME ASSEMBLY | Author keyword | 23 | 55% | 2% | 29 |
| 10 | H33 | Author keyword | 19 | 68% | 1% | 17 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | HISTONE CHAPERONE | 70 | 67% | 4% | 64 | Search HISTONE+CHAPERONE | Search HISTONE+CHAPERONE |
| 2 | ASF1 | 53 | 85% | 2% | 28 | Search ASF1 | Search ASF1 |
| 3 | CAF 1 | 32 | 78% | 1% | 21 | Search CAF+1 | Search CAF+1 |
| 4 | MACROH2A | 30 | 79% | 1% | 19 | Search MACROH2A | Search MACROH2A |
| 5 | CHROMATIN ASSEMBLY | 29 | 51% | 3% | 41 | Search CHROMATIN+ASSEMBLY | Search CHROMATIN+ASSEMBLY |
| 6 | HISTONE VARIANT | 25 | 47% | 3% | 40 | Search HISTONE+VARIANT | Search HISTONE+VARIANT |
| 7 | HISTONE VARIANTS | 25 | 44% | 3% | 43 | Search HISTONE+VARIANTS | Search HISTONE+VARIANTS |
| 8 | H2AZ | 25 | 49% | 2% | 37 | Search H2AZ | Search H2AZ |
| 9 | NUCLEOSOME ASSEMBLY | 23 | 55% | 2% | 29 | Search NUCLEOSOME+ASSEMBLY | Search NUCLEOSOME+ASSEMBLY |
| 10 | H33 | 19 | 68% | 1% | 17 | Search H33 | Search H33 |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | VARIANT H33 | 73 | 72% | 4% | 58 |
| 2 | ASF1 | 63 | 81% | 3% | 38 |
| 3 | H3 LYSINE 56 ACETYLATION | 62 | 72% | 3% | 49 |
| 4 | NEWLY SYNTHESIZED HISTONES | 54 | 64% | 4% | 54 |
| 5 | ASSEMBLY FACTOR ASF1 | 52 | 100% | 1% | 18 |
| 6 | H3K56 ACETYLATION | 52 | 75% | 3% | 38 |
| 7 | HUMAN ASF1 | 48 | 91% | 1% | 20 |
| 8 | ASSEMBLY FACTOR I | 43 | 41% | 5% | 82 |
| 9 | DEPOSITION PROTEIN ASF1 | 38 | 89% | 1% | 17 |
| 10 | DNA REPLICATION INVITRO | 38 | 44% | 4% | 65 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Histone exchange, chromatin structure and the regulation of transcription | 2015 | 7 | 155 | 61% |
| Histone variants: dynamic punctuation in transcription | 2014 | 29 | 119 | 83% |
| Histone chaperones in nucleosome assembly and human disease | 2013 | 51 | 114 | 73% |
| Every amino acid matters: essential contributions of histone variants to mammalian development and disease | 2014 | 31 | 150 | 50% |
| The double face of the histone variant H3.3 | 2011 | 78 | 90 | 68% |
| Histone Chaperones: Modulators of Chromatin Marks | 2011 | 71 | 121 | 73% |
| Histone variants - ancient wrap artists of the epigenome | 2010 | 264 | 144 | 45% |
| Histone core modifications regulating nucleosome structure and dynamics | 2014 | 19 | 54 | 56% |
| Chaperoning Histones during DNA Replication and Repair | 2010 | 141 | 99 | 66% |
| Chromatin challenges during DNA replication and repair | 2007 | 339 | 110 | 41% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | UMR 218 | 9 | 26% | 1.9% | 29 |
| 2 | UMR218 | 9 | 37% | 1.3% | 19 |
| 3 | NUCL DYNAM GENOME PLAST | 6 | 47% | 0.6% | 9 |
| 4 | DONNER 351 | 6 | 100% | 0.3% | 4 |
| 5 | CHROMATIN TRANSCRIPT REGULAT GRP | 4 | 75% | 0.2% | 3 |
| 6 | HORIKOSHI GENE SELECTOR PROJECT | 4 | 29% | 0.8% | 12 |
| 7 | CHROMATIN BIOL EPIGENET | 4 | 22% | 0.9% | 14 |
| 8 | BUTENANDT | 3 | 50% | 0.3% | 4 |
| 9 | LMU BIOMED | 3 | 60% | 0.2% | 3 |
| 10 | BIOCHEM NUCL ACIDS ENZYMOL | 2 | 67% | 0.1% | 2 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000147541 | SWI SNF//BRG1//ISWI |
| 2 | 0.0000144658 | NUCLEOSOME POSITIONING//NUCLEOSOME//NUCLEOSOME MAPPING |
| 3 | 0.0000138294 | PP32//SET TAF I BETA//INHAT |
| 4 | 0.0000136042 | SIR COMPLEX//SIR3//SIR PROTEINS |
| 5 | 0.0000127653 | LINKER HISTONE//HISTONE H1//NUCLEOSOME |
| 6 | 0.0000117551 | HISTONE DEMETHYLASE//HISTONE DEMETHYLATION//LSD1 |
| 7 | 0.0000117356 | MYST//MOZ//HISTONE ACETYLTRANSFERASE |
| 8 | 0.0000093306 | HISTONE GENE EXPRESSION//HISTONE MRNA//SLBP |
| 9 | 0.0000078987 | CENTROMERE//CENP A//NEOCENTROMERE |
| 10 | 0.0000076237 | HP1//POSITION EFFECT VARIEGATION//HETEROCHROMATIN |