Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
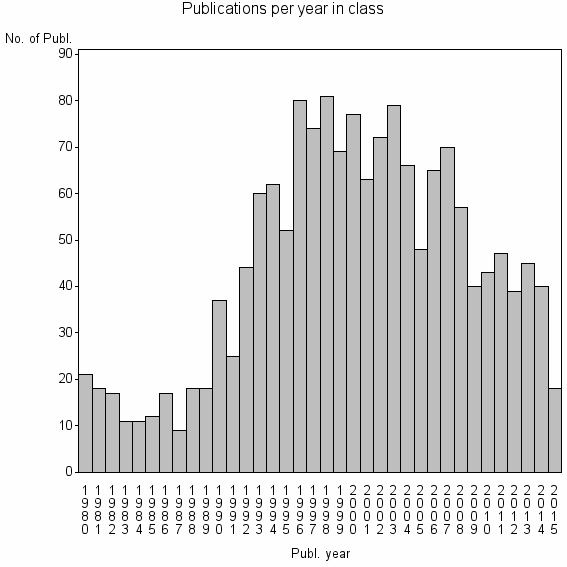
| 5100 | 1605 | 49.0 | 82% |
Classes in level above (level 2) |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | RNA DIMERIZATION | Author keyword | 41 | 87% | 1% | 20 |
| 2 | NCP7 | Author keyword | 32 | 78% | 1% | 21 |
| 3 | HIV 1 NUCLEOCAPSID PROTEIN | Author keyword | 23 | 100% | 1% | 10 |
| 4 | MCGILL AIDS | Address | 20 | 31% | 3% | 55 |
| 5 | NUCLEOCAPSID PROTEIN | Author keyword | 20 | 24% | 5% | 74 |
| 6 | NUCLEIC ACID CHAPERONE | Author keyword | 14 | 57% | 1% | 16 |
| 7 | INSERM 758 | Address | 12 | 86% | 0% | 6 |
| 8 | ORETRO | Address | 12 | 40% | 1% | 23 |
| 9 | AIDS VACCINE PROGRAM | Address | 11 | 19% | 3% | 51 |
| 10 | REVERSE TRANSCRIPTION | Author keyword | 11 | 13% | 5% | 75 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | RNA DIMERIZATION | 41 | 87% | 1% | 20 | Search RNA+DIMERIZATION | Search RNA+DIMERIZATION |
| 2 | NCP7 | 32 | 78% | 1% | 21 | Search NCP7 | Search NCP7 |
| 3 | HIV 1 NUCLEOCAPSID PROTEIN | 23 | 100% | 1% | 10 | Search HIV+1+NUCLEOCAPSID+PROTEIN | Search HIV+1+NUCLEOCAPSID+PROTEIN |
| 4 | NUCLEOCAPSID PROTEIN | 20 | 24% | 5% | 74 | Search NUCLEOCAPSID+PROTEIN | Search NUCLEOCAPSID+PROTEIN |
| 5 | NUCLEIC ACID CHAPERONE | 14 | 57% | 1% | 16 | Search NUCLEIC+ACID+CHAPERONE | Search NUCLEIC+ACID+CHAPERONE |
| 6 | REVERSE TRANSCRIPTION | 11 | 13% | 5% | 75 | Search REVERSE+TRANSCRIPTION | Search REVERSE+TRANSCRIPTION |
| 7 | TRNA3LYS | 11 | 78% | 0% | 7 | Search TRNA3LYS | Search TRNA3LYS |
| 8 | PRIMER BINDING SITE | 10 | 50% | 1% | 15 | Search PRIMER+BINDING+SITE | Search PRIMER+BINDING+SITE |
| 9 | NUCLEIC ACID CHAPERONE ACTIVITY | 8 | 100% | 0% | 5 | Search NUCLEIC+ACID+CHAPERONE+ACTIVITY | Search NUCLEIC+ACID+CHAPERONE+ACTIVITY |
| 10 | RNA PACKAGING | 8 | 35% | 1% | 18 | Search RNA+PACKAGING | Search RNA+PACKAGING |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | PRIMER BINDING SITE | 127 | 64% | 8% | 123 |
| 2 | MAJOR SPLICE DONOR | 107 | 90% | 3% | 46 |
| 3 | ZINC FINGER STRUCTURES | 102 | 88% | 3% | 49 |
| 4 | PRIMER TRANSFER RNA | 100 | 76% | 4% | 71 |
| 5 | ACID CHAPERONE ACTIVITY | 96 | 69% | 5% | 83 |
| 6 | GENOMIC RNA | 94 | 27% | 18% | 295 |
| 7 | STRONG STOP DNA | 92 | 84% | 3% | 51 |
| 8 | TRNA3LYS | 91 | 83% | 3% | 52 |
| 9 | EFFICIENT INITIATION | 86 | 90% | 2% | 37 |
| 10 | STABLE DIMER | 86 | 96% | 2% | 26 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Structural Determinants and Mechanism of HIV-1 Genome Packaging | 2011 | 61 | 253 | 73% |
| How retroviruses select their genomes | 2005 | 151 | 154 | 86% |
| Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: Critical role in reverse transcription and molecular mechanism | 2005 | 209 | 369 | 61% |
| Flexible Nature and Specific Functions of the HIV-1 Nucleocapsid Protein | 2011 | 39 | 153 | 68% |
| Dimerization of retroviral RNA genomes: An inseparable pair | 2004 | 163 | 144 | 77% |
| Nucleic-acid-chaperone activity of retroviral nucleocapsid proteins: significance for viral replication | 1998 | 279 | 50 | 80% |
| NMR structure of the HIV-1 nucleocapsid protein bound to stem-loop SL2 of the Psi-RNA packaging signal. Implications for genome recognition | 2000 | 214 | 98 | 54% |
| Strand transfer events during HIV-1 reverse transcription | 2008 | 49 | 157 | 62% |
| RNA packaging | 1996 | 268 | 168 | 86% |
| The Remarkable Frequency of Human Immunodeficiency Virus Type 1 Genetic Recombination | 2009 | 62 | 340 | 45% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | MCGILL AIDS | 20 | 31% | 3.4% | 55 |
| 2 | INSERM 758 | 12 | 86% | 0.4% | 6 |
| 3 | ORETRO | 12 | 40% | 1.4% | 23 |
| 4 | AIDS VACCINE PROGRAM | 11 | 19% | 3.2% | 51 |
| 5 | INSERM 412 | 10 | 73% | 0.5% | 8 |
| 6 | HIV DRUG ISTANCE PROGRAM | 9 | 12% | 4.3% | 69 |
| 7 | UER SCI PHARMACEUT BIOL | 6 | 80% | 0.2% | 4 |
| 8 | PROGRAM STRUCT BIOL BIOCHEM BIOPHYS | 6 | 71% | 0.3% | 5 |
| 9 | PHARMACOCHIM MOL STRUCT | 5 | 19% | 1.5% | 24 |
| 10 | U266 | 4 | 32% | 0.7% | 11 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000163746 | VIRUS CELL INTERACT SECT//GAG//M PMV |
| 2 | 0.0000115967 | NNRTIS//REVERSE TRANSCRIPTASE//HIV 1 REVERSE TRANSCRIPTASE |
| 3 | 0.0000080060 | REV//RRE//REV PROTEIN |
| 4 | 0.0000077070 | TY1//SECT EUKARYOT TRANSPOSABLE ELEMENTS//TY ELEMENTS |
| 5 | 0.0000060053 | APOBEC3G//VIF//APOBEC3F |
| 6 | 0.0000058529 | UNITE RETROVIROL MOL//ENTROPY EVOLUTION RATE//HIV 1 MOTHER TO CHILD TRANSMISSION |
| 7 | 0.0000054690 | TAT//TAT PROTEIN//TAR RNA |
| 8 | 0.0000053317 | RETROVIRAL VECTORS//RETROVIRAL VECTOR//SELECTIVE AMPLIFIER GENE |
| 9 | 0.0000052893 | AVIAN LEUKOSIS VIRUS//AVIAN LEUKOSIS VIRUS SUBGROUP J//MYELOID LEUKOSIS |
| 10 | 0.0000052336 | ARTIFICIAL TRANSCRIPTION FACTOR//ARTIFICIAL TRANSCRIPTION FACTORS//RECOGNITION CODE |